In-Depth Molecular Characterization of Neovascular Membranes Suggests a Role for Hyalocyte-to-Myofibroblast Transdifferentiation in Proliferative Diabetic Retinopathy
- PMID: 34795670
- PMCID: PMC8593213
- DOI: 10.3389/fimmu.2021.757607
In-Depth Molecular Characterization of Neovascular Membranes Suggests a Role for Hyalocyte-to-Myofibroblast Transdifferentiation in Proliferative Diabetic Retinopathy
Abstract
Background: Retinal neovascularization (RNV) membranes can lead to a tractional retinal detachment, the primary reason for severe vision loss in end-stage disease proliferative diabetic retinopathy (PDR). The aim of this study was to characterize the molecular, cellular and immunological features of RNV in order to unravel potential novel drug treatments for PDR.
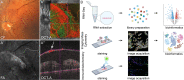
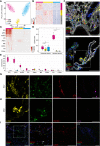
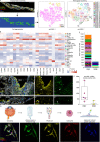
Methods: A total of 43 patients undergoing vitrectomy for PDR, macular pucker or macular hole (control patients) were included in this study. The surgically removed RNV and epiretinal membranes were analyzed by RNA sequencing, single-cell based Imaging Mass Cytometry and conventional immunohistochemistry. Immune cells of the vitreous body, also known as hyalocytes, were isolated from patients with PDR by flow cytometry, cultivated and characterized by immunohistochemistry. A bioinformatical drug repurposing approach was applied in order to identify novel potential drug options for end-stage diabetic retinopathy disease.
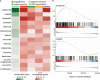
Results: The in-depth transcriptional and single-cell protein analysis of diabetic RNV tissue samples revealed an accumulation of endothelial cells, macrophages and myofibroblasts as well as an abundance of secreted ECM proteins such as SPARC, FN1 and several types of collagen in RNV tissue. The immunohistochemical staining of cultivated vitreal hyalocytes from patients with PDR showed that hyalocytes express α-SMA (alpha-smooth muscle actin), a classic myofibroblast marker. According to our drug repurposing analysis, imatinib emerged as a potential immunomodulatory drug option for future treatment of PDR.
Conclusion: This study delivers the first in-depth transcriptional and single-cell proteomic characterization of RNV tissue samples. Our data suggest an important role of hyalocyte-to-myofibroblast transdifferentiation in the pathogenesis of diabetic vitreoretinal disease and their modulation as a novel possible clinical approach.
Keywords: Imaging Mass Cytometry; RNA sequencing; hyalocytes; myofibroblasts; proliferative diabetic retinopathy (PDR); retinal neovascularization (RNV); transdifferentiation.
Copyright © 2021 Boneva, Wolf, Hajdú, Prinz, Salié, Schlecht, Killmer, Laich, Faatz, Lommatzsch, Busch, Bucher, Stahl, Böhringer, Bengsch, Schlunck, Agostini and Lange.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

