Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb
- PMID: 34798069
- PMCID: PMC8564600
- DOI: 10.1016/j.cell.2021.10.027
Visualizing in deceased COVID-19 patients how SARS-CoV-2 attacks the respiratory and olfactory mucosae but spares the olfactory bulb
Abstract
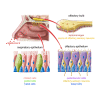
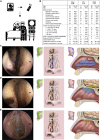
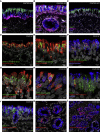
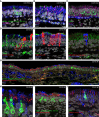
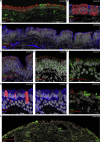
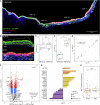
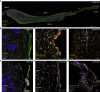
Anosmia, the loss of smell, is a common and often the sole symptom of COVID-19. The onset of the sequence of pathobiological events leading to olfactory dysfunction remains obscure. Here, we have developed a postmortem bedside surgical procedure to harvest endoscopically samples of respiratory and olfactory mucosae and whole olfactory bulbs. Our cohort of 85 cases included COVID-19 patients who died a few days after infection with SARS-CoV-2, enabling us to catch the virus while it was still replicating. We found that sustentacular cells are the major target cell type in the olfactory mucosa. We failed to find evidence for infection of olfactory sensory neurons, and the parenchyma of the olfactory bulb is spared as well. Thus, SARS-CoV-2 does not appear to be a neurotropic virus. We postulate that transient insufficient support from sustentacular cells triggers transient olfactory dysfunction in COVID-19. Olfactory sensory neurons would become affected without getting infected.
Keywords: B.1.1.7; COVID-19; SARS-CoV-2; UGT2A1; coronavirus; leptomeninges; olfactory bulb; olfactory receptor; olfactory sensory neuron; sustentacular cell.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.D.H., A.N., L.P., and J.W.R. are employees and stockholders at NanoString Technologies, Inc.
Figures






References
-
- Acevedo C., Blanchard K., Bacigalupo J., Vergara C. Possible ATP trafficking by ATP-shuttles in the olfactory cilia and glucose transfer across the olfactory mucosa. FEBS Lett. 2019;593:601–610. - PubMed
-
- Barnes I.H.A., Ibarra-Soria X., Fitzgerald S., Gonzalez J.M., Davidson C., Hardy M.P., Manthravadi D., Van Gerven L., Jorissen M., Zeng Z., et al. Expert curation of the human and mouse olfactory receptor gene repertoires identifies conserved coding regions split across two exons. BMC Genomics. 2020;21:196. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

