MhcVizPipe: A Quality Control Software for Rapid Assessment of Small- to Large-Scale Immunopeptidome Datasets
- PMID: 34798331
- PMCID: PMC8717601
- DOI: 10.1016/j.mcpro.2021.100178
MhcVizPipe: A Quality Control Software for Rapid Assessment of Small- to Large-Scale Immunopeptidome Datasets
Abstract
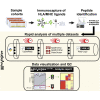
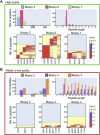
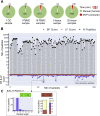
MS-based immunopeptidomics is maturing into an automatized and high-throughput technology, producing small- to large-scale datasets of clinically relevant major histocompatibility complex (MHC) class I-associated and class II-associated peptides. Consequently, the development of quality control (QC) and quality assurance systems capable of detecting sample and/or measurement issues is important for instrument operators and scientists in charge of downstream data interpretation. Here, we created MhcVizPipe (MVP), a semiautomated QC software tool that enables rapid and simultaneous assessment of multiple MHC class I and II immunopeptidomic datasets generated by MS, including datasets generated from large sample cohorts. In essence, MVP provides a rapid and consolidated view of sample quality, composition, and MHC specificity to greatly accelerate the "pass-fail" QC decision-making process toward data interpretation. MVP parallelizes the use of well-established immunopeptidomic algorithms (NetMHCpan, NetMHCIIpan, and GibbsCluster) and rapidly generates organized and easy-to-understand reports in HTML format. The reports are fully portable and can be viewed on any computer with a modern web browser. MVP is intuitive to use and will find utility in any specialized immunopeptidomic laboratory and proteomics core facility that provides immunopeptidomic services to the community.
Keywords: MHC; MS; immunopeptidomics; peptide; software.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest A. J. and E. P. are employees of CellCarta (Montreal, Canada); M. T., L. R., and R. B. are employees of Biognosys (Zürich, Switzerland). All other authors declare no competing interests.
Figures


References
-
- Gonzalez-Duque S., Azoury M.E., Colli M.L., Afonso G., Turatsinze J.-V., Nigi L., Lalanne A.I., Sebastiani G., Carré A., Pinto S., Culina S., Corcos N., Bugliani M., Marchetti P., Armanet M., et al. Conventional and neo-antigenic peptides presented by β cells are targeted by circulating NaIve CD8+ T cells in type 1 diabetic and healthy donors. Cell Metab. 2018;28:946–960.e6. - PubMed
-
- Gubin M.M., Zhang X., Schuster H., Caron E., Ward J.P., Noguchi T., Ivanova Y., Hundal J., Arthur C.D., Krebber W.-J., Mulder G.E., Toebes M., Vesely M.D., Lam S.S.K., Korman A.J., et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515:577–581. - PMC - PubMed
-
- Bassani-Sternberg M., Bräunlein E., Klar R., Engleitner T., Sinitcyn P., Audehm S., Straub M., Weber J., Slotta-Huspenina J., Specht K., Martignoni M.E., Werner A., Hein R., Busch D.H., Peschel C., et al. Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat. Commun. 2016;7:13404. - PMC - PubMed
-
- Yadav M., Jhunjhunwala S., Phung Q.T., Lupardus P., Tanguay J., Bumbaca S., Franci C., Cheung T.K., Fritsche J., Weinschenk T., Modrusan Z., Mellman I., Lill J.R., Delamarre L. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature. 2015;515:572–576. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

