Perspectives on SARS-CoV-2 Main Protease Inhibitors
- PMID: 34798775
- PMCID: PMC9357291
- DOI: 10.1021/acs.jmedchem.1c00409
Perspectives on SARS-CoV-2 Main Protease Inhibitors
Abstract
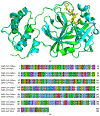
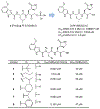
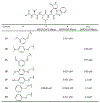
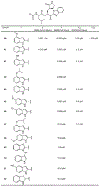
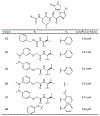
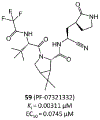
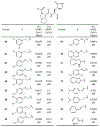
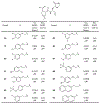
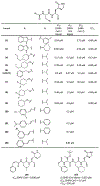
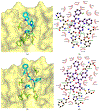
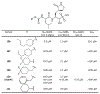
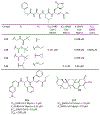
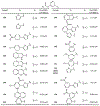
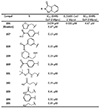
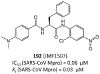
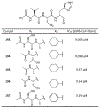
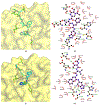
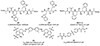
The main protease (Mpro) plays a crucial role in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) replication and is highly conserved, rendering it one of the most attractive therapeutic targets for SARS-CoV-2 inhibition. Currently, although two drug candidates targeting SARS-CoV-2 Mpro designed by Pfizer are under clinical trials, no SARS-CoV-2 medication is approved due to the long period of drug development. Here, we collect a comprehensive list of 817 available SARS-CoV-2 and SARS-CoV Mpro inhibitors from the literature or databases and analyze their molecular mechanisms of action. The structure-activity relationships (SARs) among each series of inhibitors are discussed. Additionally, we broadly examine available antiviral activity, ADMET (absorption, distribution, metabolism, excretion, and toxicity), and animal tests of these inhibitors. We comment on their druggability or drawbacks that prevent them from becoming drugs. This Perspective sheds light on the future development of Mpro inhibitors for SARS-CoV-2 and future coronavirus diseases.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

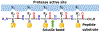
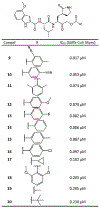
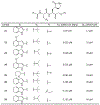
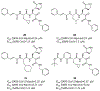


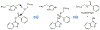
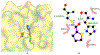

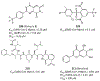
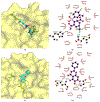
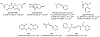
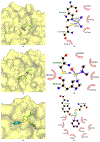
References
-
- Shin MD; Shukla S; Chung YH; Beiss V; Chan SK; Ortega-Rivera OA; Wirth DM; Chen A; Sack M; Pokorski JK; Steinmetz NF COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol 2020, 15, 646–655. - PubMed
-
- Zimmer C; Corum J; Wee S-L Coronavirus Vaccine Tracker. https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tra..., 2020.
-
- Halford B Pfizer unveils its oral SARS-CoV-2 inhibitor. https://cen.acs.org/acs-news/acs-meeting-news/Pfizer-unveils-oral-SARS-C..., 2020.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

