Self-organization of human dorsal-ventral forebrain structures by light induced SHH
- PMID: 34799555
- PMCID: PMC8604999
- DOI: 10.1038/s41467-021-26881-w
Self-organization of human dorsal-ventral forebrain structures by light induced SHH
Abstract
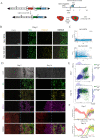
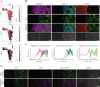
Organizing centers secrete morphogens that specify the emergence of germ layers and the establishment of the body's axes during embryogenesis. While traditional experimental embryology tools have been instrumental in dissecting the molecular aspects of organizers in model systems, they are impractical in human in-vitro model systems to dissect the relationships between signaling and fate along embryonic coordinates. To systematically study human embryonic organizer centers, we devised a collection of optogenetic ePiggyBac vectors to express a photoactivatable Cre-loxP recombinase, that allows the systematic induction of organizer structures by shining blue-light on human embryonic stem cells (hESCs). We used a light stimulus to geometrically confine SHH expression in neuralizing hESCs. This led to the self-organization of mediolateral neural patterns. scRNA-seq analysis established that these structures represent the dorsal-ventral forebrain, at the end of the first month of development. Here, we show that morphogen light-stimulation is a scalable tool that induces self-organizing centers.
© 2021. The Author(s).
Conflict of interest statement
A.H.B. is the co-founder of RUMI Scientific, RUMI Viro and OvaNova. A.H.B., F.E., and E.A.R. are shareholders of RUMI Scientific and RUMI Viro. R.D.S. has no competing interests to disclose.
Figures


References
-
- Spemann H, Mangold H. Induction of embryonic primordia by implantation of organizers from a different species. 1923. Int J. Dev. Biol. 2001;45:13–38. - PubMed
-
- Harland R, Gerhart J. Formation and function of spemann’s organizer. Cell Dev. Biol. 1997;13:611–667. - PubMed
-
- Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77:273–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

