Protein innovation through template switching in the Saccharomyces cerevisiae lineage
- PMID: 34799587
- PMCID: PMC8604942
- DOI: 10.1038/s41598-021-01736-y
Protein innovation through template switching in the Saccharomyces cerevisiae lineage
Abstract
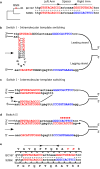
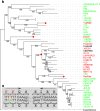
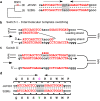
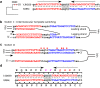
DNA polymerase template switching between short, non-identical inverted repeats (IRs) is a genetic mechanism that leads to the homogenization of IR arms and to IR spacer inversion, which cause multinucleotide mutations (MNMs). It is unknown if and how template switching affects gene evolution. In this study, we performed a phylogenetic analysis to determine the effect of template switching between IR arms on coding DNA of Saccharomyces cerevisiae. To achieve this, perfect IRs that co-occurred with MNMs between a strain and its parental node were identified in S. cerevisiae strains. We determined that template switching introduced MNMs into 39 protein-coding genes through S. cerevisiae evolution, resulting in both arm homogenization and inversion of the IR spacer. These events in turn resulted in nonsynonymous substitutions and up to five neighboring amino acid replacements in a single gene. The study demonstrates that template switching is a powerful generator of multiple substitutions within codons. Additionally, some template switching events occurred more than once during S. cerevisiae evolution. Our findings suggest that template switching constitutes a general mutagenic mechanism that results in both nonsynonymous substitutions and parallel evolution, which are traditionally considered as evidence for positive selection, without the need for adaptive explanations.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

