Allelic variants of full-length VAR2CSA, the placental malaria vaccine candidate, differ in antigenicity and receptor binding affinity
- PMID: 34799664
- PMCID: PMC8604988
- DOI: 10.1038/s42003-021-02787-7
Allelic variants of full-length VAR2CSA, the placental malaria vaccine candidate, differ in antigenicity and receptor binding affinity
Erratum in
-
Author Correction: Allelic variants of full-length VAR2CSA, the placental malaria vaccine candidate, differ in antigenicity and receptor binding affinity.Commun Biol. 2022 Apr 26;5(1):404. doi: 10.1038/s42003-022-03337-5. Commun Biol. 2022. PMID: 35474125 Free PMC article. No abstract available.
Abstract
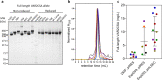
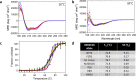
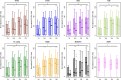
Plasmodium falciparum-infected erythrocytes (IE) sequester in the placenta via surface protein VAR2CSA, which binds chondroitin sulfate A (CSA) expressed on the syncytiotrophoblast surface, causing placental malaria (PM) and severe adverse outcomes in mothers and their offspring. VAR2CSA belongs to the PfEMP1 variant surface antigen family; PfEMP1 proteins mediate IE adhesion and facilitate parasite immunoevasion through antigenic variation. Here we produced deglycosylated (native-like) and glycosylated versions of seven recombinant full-length VAR2CSA ectodomains and compared them for antigenicity and adhesiveness. All VAR2CSA recombinants bound CSA with nanomolar affinity, and plasma from Malian pregnant women demonstrated antigen-specific reactivity that increased with gravidity and trimester. However, allelic and glycosylation variants differed in their affinity to CSA and their serum reactivities. Deglycosylated proteins (native-like) showed higher CSA affinity than glycosylated proteins for all variants except NF54. Further, the gravidity-related increase in serum VAR2CSA reactivity (correlates with acquisition of protective immunity) was absent with the deglycosylated form of atypical M200101 VAR2CSA with an extended C-terminal region. Our findings indicate significant inter-allelic differences in adhesion and seroreactivity that may contribute to the heterogeneity of clinical presentations, which could have implications for vaccine design.
© 2021. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests as defined by Nature Research, or other interests that might be perceived to influence the results and/or discussion reported in this paper.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

