Manifold learning analysis suggests strategies to align single-cell multimodal data of neuronal electrophysiology and transcriptomics
- PMID: 34799674
- PMCID: PMC8604989
- DOI: 10.1038/s42003-021-02807-6
Manifold learning analysis suggests strategies to align single-cell multimodal data of neuronal electrophysiology and transcriptomics
Abstract
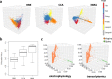
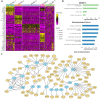
Recent single-cell multimodal data reveal multi-scale characteristics of single cells, such as transcriptomics, morphology, and electrophysiology. However, integrating and analyzing such multimodal data to deeper understand functional genomics and gene regulation in various cellular characteristics remains elusive. To address this, we applied and benchmarked multiple machine learning methods to align gene expression and electrophysiological data of single neuronal cells in the mouse brain from the Brain Initiative. We found that nonlinear manifold learning outperforms other methods. After manifold alignment, the cells form clusters highly corresponding to transcriptomic and morphological cell types, suggesting a strong nonlinear relationship between gene expression and electrophysiology at the cell-type level. Also, the electrophysiological features are highly predictable by gene expression on the latent space from manifold alignment. The aligned cells further show continuous changes of electrophysiological features, implying cross-cluster gene expression transitions. Functional enrichment and gene regulatory network analyses for those cell clusters revealed potential genome functions and molecular mechanisms from gene expression to neuronal electrophysiology.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

