Dcf1 induces glioblastoma cells apoptosis by blocking autophagy
- PMID: 34799992
- PMCID: PMC8704163
- DOI: 10.1002/cam4.4440
Dcf1 induces glioblastoma cells apoptosis by blocking autophagy
Abstract
Background: Dcf1 has been demonstrated to play vital roles in many CNS diseases, it also has a destructive role on cell mitochondria in glioma cells and promotes the autophagy. Hitherto, it is unclear whether the viability of glioblastoma cells is affected by Dcf1, in particular Dcf1 possesses broad localization on different organelles, and the organelles interaction frequently implicated in cancer cells survival.
Methods: Surgically excised WHO grade IV human glioblastoma tissues were collected and cells isolated for culturing. RT-PCR and DNA sequencing assay to estimate the abundance and mutation of Dcf1. iTRAQ sequencing and bioinformatic analysis were performed. Subsequently, immunoprecipitation assay to evaluate the degradation of HistoneH2A isomers by UBA52 ubiquitylation. Transmission electron microscopy (TEM) was applied to observe the structure change of mitochondria and autophagosome. Organelle isolated assay to determine the distribution of protein. Cell cycle and apoptosis were evaluated by flow cytometric assays.
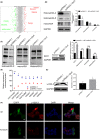
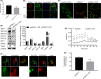
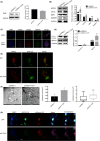
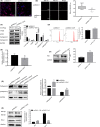
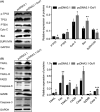
Results: Dcf1 was downregulated in WHO grade IV tumor without mutation, and overexpression of Dcf1 was found to significantly regulate glioblastoma cells. One hundred and seventy-six differentially expressed proteins were identified by iTRAQ sequencing. Furthermore, we confirmed that overexpression of Dcf1 destabilized the structure of the nucleosome via UBA52 ubiquitination to downregulate HistoneH2A.X but not macroH2A or HistoneH2A.Z, decreased the mitochondrial DNA copy number and inhibited the mitochondrial biogenesis, thus causing mitochondrial destruction and dysfunction in order to supply cellular energy and induce mitophagy preferentially but not apoptosis. Dcf1 also has disrupted the integrity of lysosomes to block autolysosome degradation and autophagy and to increase the release of Cathepsin B and D from lysosomes into cytosol. These proteins cleaved and activated BID to induce glioblastoma cells apoptosis.
Conclusions: In this study, we demonstrated that unmutated Dcf1 expression is negatively related to the malignancy of glioblastoma, Dcf1 overexpression causes nucleosomes destabilization, mitochondria destruction and dysfunction to induce mitophagy preferentially, and block autophagy by impairing lysosomes to induce apoptosis in glioblastoma.
Keywords: Dcf1; apoptosis; glioblastoma; lysosome; mitochondria; mitophagy.
© 2021 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Figures
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7‐33. - PubMed
-
- Lim M, Xia YX, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol. 2018;15:422‐442. - PubMed
-
- Di Mascolo D, Palange AL, Primavera R, et al. Conformable hierarchically engineered polymeric micromeshes enabling combinatorial therapies in brain tumours. Nat Nanotechnol. 2021;16(7):820‐829. - PubMed
-
- Zou Y, Liu YJ, Yang ZP, Zhang DY, Lu YQ, Zheng M, Xue X, Geng J, Chung R, Shi BY. Effective and targeted human orthotopic glioblastoma xenograft therapy via a multifunctional biomimetic nanomedicine. Adv Mater. 2018;30(51):1803717. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

