Comparative analysis of transposable elements provides insights into genome evolution in the genus Camelus
- PMID: 34800971
- PMCID: PMC8605555
- DOI: 10.1186/s12864-021-08117-9
Comparative analysis of transposable elements provides insights into genome evolution in the genus Camelus
Abstract
Background: Transposable elements (TEs) are common features in eukaryotic genomes that are known to affect genome evolution critically and to play roles in gene regulation. Vertebrate genomes are dominated by TEs, which can reach copy numbers in the hundreds of thousands. To date, details regarding the presence and characteristics of TEs in camelid genomes have not been made available.
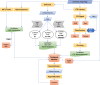
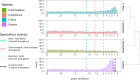
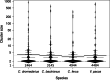
Results: We conducted a genome-wide comparative analysis of camelid TEs, focusing on the identification of TEs and elucidation of transposition histories in four species: Camelus dromedarius, C. bactrianus, C. ferus, and Vicugna pacos. Our TE library was created using both de novo structure-based and homology-based searching strategies ( https://github.com/kacst-bioinfo-lab/TE_ideintification_pipeline ). Annotation results indicated a similar proportion of each genomes comprising TEs (35-36%). Class I LTR retrotransposons comprised 16-20% of genomes, and mostly consisted of the endogenous retroviruses (ERVs) groups ERVL, ERVL-MaLR, ERV_classI, and ERV_classII. Non-LTR elements comprised about 12% of genomes and consisted of SINEs (MIRs) and the LINE superfamilies LINE1, LINE2, L3/CR1, and RTE clades. Least represented were the Class II DNA transposons (2%), consisting of hAT-Charlie, TcMar-Tigger, and Helitron elements and comprising about 1-2% of each genome.
Conclusions: The findings of the present study revealed that the distribution of transposable elements across camelid genomes is approximately similar. This investigation presents a characterization of TE content in four camelid to contribute to developing a better understanding of camelid genome architecture and evolution.
Keywords: Camelid genomes; De novo TEs annotation; Retrotransposons; Transposable elements; Transposons.
© 2021. The Author(s).
Conflict of interest statement
The authors declare there are no competing interests.
Figures




Similar articles
-
Genome-wide comparative analysis of transposable elements in Palmae genomes.Front Biosci (Landmark Ed). 2021 Nov 30;26(11):1119-1131. doi: 10.52586/5014. Front Biosci (Landmark Ed). 2021. PMID: 34856758
-
Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates.Genome Biol Evol. 2015 Jan 9;7(2):567-80. doi: 10.1093/gbe/evv005. Genome Biol Evol. 2015. PMID: 25577199 Free PMC article.
-
Evolutionary active transposable elements in the genome of the coelacanth.J Exp Zool B Mol Dev Evol. 2014 Sep;322(6):322-33. doi: 10.1002/jez.b.22521. Epub 2013 Aug 1. J Exp Zool B Mol Dev Evol. 2014. PMID: 23908136
-
Mammalian transposable elements and their impacts on genome evolution.Chromosome Res. 2018 Mar;26(1-2):25-43. doi: 10.1007/s10577-017-9570-z. Epub 2018 Feb 1. Chromosome Res. 2018. PMID: 29392473 Free PMC article. Review.
-
Structural and sequence diversity of eukaryotic transposable elements.Genes Genet Syst. 2020 Jan 30;94(6):233-252. doi: 10.1266/ggs.18-00024. Epub 2018 Nov 9. Genes Genet Syst. 2020. PMID: 30416149 Review.
Cited by
-
Mitochondrial DNA of the Arabian Camel Camelus dromedarius.Animals (Basel). 2024 Aug 24;14(17):2460. doi: 10.3390/ani14172460. Animals (Basel). 2024. PMID: 39272245 Free PMC article. Review.
-
Taming transposable elements in livestock and poultry: a review of their roles and applications.Genet Sel Evol. 2023 Jul 21;55(1):50. doi: 10.1186/s12711-023-00821-2. Genet Sel Evol. 2023. PMID: 37479995 Free PMC article. Review.
-
LTR Retroelements and Bird Adaptation to Arid Environments.Int J Mol Sci. 2023 Mar 28;24(7):6332. doi: 10.3390/ijms24076332. Int J Mol Sci. 2023. PMID: 37047324 Free PMC article.
-
High Diversity of Long Terminal Repeat Retrotransposons in Compact Vertebrate Genomes: Insights from Genomes of Tetraodontiformes.Animals (Basel). 2024 May 10;14(10):1425. doi: 10.3390/ani14101425. Animals (Basel). 2024. PMID: 38791643 Free PMC article.
-
Comparative genomics reveals insights into anuran genome size evolution.BMC Genomics. 2023 Jul 6;24(1):379. doi: 10.1186/s12864-023-09499-8. BMC Genomics. 2023. PMID: 37415107 Free PMC article.
References
-
- Adelson D, Raison J, Garber M, Edgar R. Interspersed repeats in the horse (equus caballus); spatial correlations highlight conserved chromosomal domains. Anim Genet. 2010;41:91–9. - PubMed
-
- Al-Swailem AM, Shehata MM, Abu-Duhier FM, Al-Yamani EJ, Al-Busadah KA, Al-Arawi MS, Al-Khider AY, Al-Muhaimeed AN, Al-Qahtani FH, Manee MM, Al-Shomrani BM, Al-Qhtani SM, Al-Harthi AS, Akdemir KC, Inan MS, Otu HH. Sequencing, Analysis, and Annotation of Expressed Sequence Tags for Camelus dromedarius. PLoS ONE. 2010;5:e10720. - PMC - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

