Development of a versatile HPLC-based method to evaluate the activation status of small GTPases
- PMID: 34801548
- PMCID: PMC8668980
- DOI: 10.1016/j.jbc.2021.101428
Development of a versatile HPLC-based method to evaluate the activation status of small GTPases
Abstract
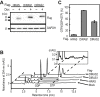
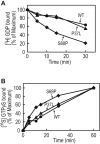
Small GTPases cycle between an inactive GDP-bound and an active GTP-bound state to control various cellular events, such as cell proliferation, cytoskeleton organization, and membrane trafficking. Clarifying the guanine nucleotide-bound states of small GTPases is vital for understanding the regulation of small GTPase functions and the subsequent cellular responses. Although several methods have been developed to analyze small GTPase activities, our knowledge of the activities for many small GTPases is limited, partly because of the lack of versatile methods to estimate small GTPase activity without unique probes and specialized equipment. In the present study, we developed a versatile and straightforward HPLC-based assay to analyze the activation status of small GTPases by directly quantifying the amounts of guanine nucleotides bound to them. This assay was validated by analyzing the RAS-subfamily GTPases, including HRAS, which showed that the ratios of GTP-bound forms were comparable with those obtained in previous studies. Furthermore, we applied this assay to the investigation of psychiatric disorder-associated mutations of RHEB (RHEB/P37L and RHEB/S68P), revealing that both mutations cause an increase in the ratio of the GTP-bound form in cells. Mechanistically, loss of sensitivity to TSC2 (a GTPase-activating protein for RHEB) for RHEB/P37L, as well as both decreased sensitivity to TSC2 and accelerated guanine-nucleotide exchange for RHEB/S68P, is involved in the increase of their GTP-bound forms, respectively. In summary, the HPLC-based assay developed in this study provides a valuable tool for analyzing small GTPases for which the activities and regulatory mechanisms are less well understood.
Keywords: GTPase-activating protein; RHEB; Ras protein; high-performance liquid chromatography; mammalian target of rapamycin; signal transduction; small GTPase; tuberous sclerosis complex.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

