Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets
- PMID: 34801852
- PMCID: PMC8585639
- DOI: 10.1016/j.biopha.2021.112420
Molecular pathways involved in COVID-19 and potential pathway-based therapeutic targets
Abstract
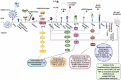
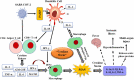
Deciphering the molecular downstream consequences of severe acute respiratory syndrome coronavirus (SARS-CoV)- 2 infection is important for a greater understanding of the disease and treatment planning. Furthermore, greater understanding of the underlying mechanisms of diagnostic and therapeutic strategies can help in the development of vaccines and drugs against COVID-19. At present, the molecular mechanisms of SARS-CoV-2 in the host cells are not sufficiently comprehended. Some of the mechanisms are proposed considering the existing similarities between SARS-CoV-2 and the other members of the β-CoVs, and others are explained based on studies advanced in the structure and function of SARS-CoV-2. In this review, we endeavored to map the possible mechanisms of the host response following SARS-CoV-2 infection and surveyed current research conducted by in vitro, in vivo and human observations, as well as existing suggestions. We addressed the specific signaling events that can cause cytokine storm and demonstrated three forms of cell death signaling following virus infection, including apoptosis, pyroptosis, and necroptosis. Given the elicited signaling pathways, we introduced possible pathway-based therapeutic targets; ADAM17 was especially highlighted as one of the most important elements of several signaling pathways involved in the immunopathogenesis of COVID-19. We also provided the possible drug candidates against these targets. Moreover, the cytokine-cytokine receptor interaction pathway was found as one of the important cross-talk pathways through a pathway-pathway interaction analysis for SARS-CoV-2 infection.
Keywords: COVID-19; Drug targets; Molecular pathway; SARS-CoV-2.
Copyright © 2021. Published by Elsevier Masson SAS.
Conflict of interest statement
The authors have no conflict of interest to report.
Figures






References
-
- Khan A., Gui J., Ahmad W., Haq I., Shahid M., Khan A.A., Shah A., Khan A., Ali L., Anwar Z., Safdar M., Abubaker J., Uddin N.N., Cao L., Wei D.Q., Mohammad A. The SARS-CoV-2 B. 1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: a computational structural perspective. RSC Adv. 2021;11(48):30132–30147. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

