On some fundamental challenges in monitoring epidemics
- PMID: 34802270
- PMCID: PMC8607144
- DOI: 10.1098/rsta.2021.0117
On some fundamental challenges in monitoring epidemics
Abstract
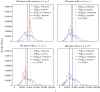
Epidemic models often reflect characteristic features of infectious spreading processes by coupled nonlinear differential equations considering different states of health (such as susceptible, infectious or recovered). This compartmental modelling approach, however, delivers an incomplete picture of the dynamics of epidemics, as it neglects stochastic and network effects, and the role of the measurement process, on which the estimation of epidemiological parameters and incidence values relies. In order to study the related issues, we combine established epidemiological spreading models with a measurement model of the testing process, considering the problems of false positives and false negatives as well as biased sampling. Studying a model-generated ground truth in conjunction with simulated observation processes (virtual measurements) allows one to gain insights into the fundamental limitations of purely data-driven methods when assessing the epidemic situation. We conclude that epidemic monitoring, simulation, and forecasting are wicked problems, as applying a conventional data-driven approach to a complex system with nonlinear dynamics, network effects and uncertainty can be misleading. Nevertheless, some of the errors can be corrected for, using scientific knowledge of the spreading dynamics and the measurement process. We conclude that such corrections should generally be part of epidemic monitoring, modelling and forecasting efforts. This article is part of the theme issue 'Data science approaches to infectious disease surveillance'.
Keywords: complex systems; computer simulation; data science; epidemic modelling; network theory; statistics.
Figures
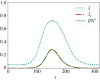
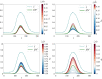
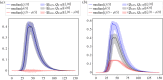
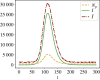
Similar articles
-
Translating surveillance data into incidence estimates.Philos Trans R Soc Lond B Biol Sci. 2019 Jul 8;374(1776):20180262. doi: 10.1098/rstb.2018.0262. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31104599 Free PMC article.
-
Epidemic Forecasting is Messier Than Weather Forecasting: The Role of Human Behavior and Internet Data Streams in Epidemic Forecast.J Infect Dis. 2016 Dec 1;214(suppl_4):S404-S408. doi: 10.1093/infdis/jiw375. J Infect Dis. 2016. PMID: 28830111 Free PMC article. Review.
-
Detection, forecasting and control of infectious disease epidemics: modelling outbreaks in humans, animals and plants.Philos Trans R Soc Lond B Biol Sci. 2019 Jun 24;374(1775):20190038. doi: 10.1098/rstb.2019.0038. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31056051 Free PMC article.
-
Susceptible-Infectious-Susceptible Epidemic Model with Symmetrical Fluctuations: Equilibrium States and Stability Analyses for Finite Systems.Acta Biotheor. 2024 Nov 13;72(4):13. doi: 10.1007/s10441-024-09490-0. Acta Biotheor. 2024. PMID: 39535581
-
Data-driven methods for present and future pandemics: Monitoring, modelling and managing.Annu Rev Control. 2021;52:448-464. doi: 10.1016/j.arcontrol.2021.05.003. Epub 2021 Jun 29. Annu Rev Control. 2021. PMID: 34220287 Free PMC article. Review.
Cited by
-
Estimation of Excess Deaths Associated With the COVID-19 Pandemic in Istanbul, Turkey.Front Public Health. 2022 Jul 25;10:888123. doi: 10.3389/fpubh.2022.888123. eCollection 2022. Front Public Health. 2022. PMID: 35958866 Free PMC article.
-
Operational analysis for COVID-19 testing: Determining the risk from asymptomatic infections.PLoS One. 2023 Feb 13;18(2):e0281710. doi: 10.1371/journal.pone.0281710. eCollection 2023. PLoS One. 2023. PMID: 36780871 Free PMC article.
-
Data science approaches to infectious disease surveillance.Philos Trans A Math Phys Eng Sci. 2022 Jan 10;380(2214):20210115. doi: 10.1098/rsta.2021.0115. Epub 2021 Nov 22. Philos Trans A Math Phys Eng Sci. 2022. PMID: 34802266 Free PMC article. Review.
References
-
- Bharadwaj KK et al. 2021. Computational intelligence in vaccine design against COVID-19. In Computational intelligence methods in COVID-19: surveillance, prevention, prediction and diagnosis, pp. 311–329. Berlin, Germany: Springer. (10.1007/978-981-15-8534-0_16) - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

