Using deep-neural-network-driven facial recognition to identify distinct Kabuki syndrome 1 and 2 gestalt
- PMID: 34803161
- PMCID: PMC9177756
- DOI: 10.1038/s41431-021-00994-8
Using deep-neural-network-driven facial recognition to identify distinct Kabuki syndrome 1 and 2 gestalt
Abstract
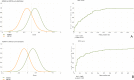
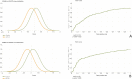
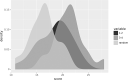
Kabuki syndrome (KS) is a rare genetic disorder caused by mutations in two major genes, KMT2D and KDM6A, that are responsible for Kabuki syndrome 1 (KS1, OMIM147920) and Kabuki syndrome 2 (KS2, OMIM300867), respectively. We lack a description of clinical signs to distinguish KS1 and KS2. We used facial morphology analysis to detect any facial morphological differences between the two KS types. We used a facial-recognition algorithm to explore any facial morphologic differences between the two types of KS. We compared several image series of KS1 and KS2 individuals, then compared images of those of Caucasian origin only (12 individuals for each gene) because this was the main ethnicity in this series. We also collected 32 images from the literature to amass a large series. We externally validated results obtained by the algorithm with evaluations by trained clinical geneticists using the same set of pictures. Use of the algorithm revealed a statistically significant difference between each group for our series of images, demonstrating a different facial morphotype between KS1 and KS2 individuals (mean area under the receiver operating characteristic curve = 0.85 [p = 0.027] between KS1 and KS2). The algorithm was better at discriminating between the two types of KS with images from our series than those from the literature (p = 0.0007). Clinical geneticists trained to distinguished KS1 and KS2 significantly recognised a unique facial morphotype, which validated algorithm findings (p = 1.6e-11). Our deep-neural-network-driven facial-recognition algorithm can reveal specific composite gestalt images for KS1 and KS2 individuals.
© 2021. The Author(s), under exclusive licence to European Society of Human Genetics.
Conflict of interest statement
DG was a consultant for the Takeda Society in 2018. Takeda did not have any role in this study.
Figures

References
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

