β-Carboline alkaloids induce structural plasticity and inhibition of SARS-CoV-2 nsp3 macrodomain more potently than remdesivir metabolite GS-441524: computational approach
- PMID: 34803450
- PMCID: PMC8573841
- DOI: 10.3906/biy-2106-64
β-Carboline alkaloids induce structural plasticity and inhibition of SARS-CoV-2 nsp3 macrodomain more potently than remdesivir metabolite GS-441524: computational approach
Abstract
The nsp3 macrodomain is implicated in the viral replication, pathogenesis and host immune responses through the removal of ADP-ribosylation sites during infections of coronaviruses including the SARS-CoV-2. It has ever been modulated by macromolecules including the ADP-ribose until Ni and co-workers recently reported its inhibition and plasticity enhancement unprecedentedly by remdesivir metabolite, GS-441524, creating an opportunity for investigating other biodiverse small molecules such as β-Carboline (βC) alkaloids. In this study, 1497 βC analogues from the HiT2LEAD chemical database were screened, using computational approaches of Glide XP docking, molecular dynamics simulation and pk-CSM ADMET predictions. Selectively, βC ligands, 129, 584, 1303 and 1323 demonstrated higher binding affinities to the receptor, indicated by XP docking scores of -10.72, -10.01, -9.63 and -9.48 kcal/mol respectively than remdesivir and GS-441524 with -4.68 and -9.41 kcal/mol respectively. Consistently, their binding free energies were -36.07, -23.77, -24.07 and -17.76 kcal/mol respectively, while remdesivir and GS-441524 showed -21.22 and -24.20 kcal/mol respectively. Interestingly, the selected βC ligands displayed better stability and flexibility for enhancing the plasticity of the receptor than GS-441524, especially 129 and 1303. Their predicted ADMET parameters favour druggability and low expressions for toxicity. Thus, they are recommended as promising adjuvant/standalone anti-SARS-CoV-2 candidates for further study.Key words: SARS-CoV-2, nsp3 macrodomain, ADP-ribose, β-carboline, bioinformatics, drug design.
Keywords: ADP-ribose; SARS-CoV-2; bioinformatics; drug design; nsp3 macrodomain; β-carboline.
Copyright © 2021 The Author(s).
Conflict of interest statement
CONFLICT OF INTEREST: The authors declare no conflict of interest.
Figures
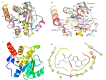


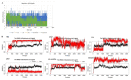

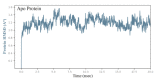



References
-
- Anderson AC Neil RHO Surti TS Stroud RM Approaches to solving the rigid receptor problem by identifying a minimal set of £ exible residues during ligand docking 1. Chemistry and Biology. 2001;8:445–457. - PubMed
-
- Ayipo YO Mordi MN Mustapha M Damodaran T Neuropharmacological potentials of β-carboline alkaloids for neuropsychiatric disorders. European Journal of Pharmacology. 2021;893:173837–173837. - PubMed
-
- Ayipo YO Osunniran WA Mordi MN Metal complexes of β-carboline: Advances in anticancer therapeutics. Coordination Chemistry Reviews. 2021;432:213746–213746.
LinkOut - more resources
Full Text Sources
Miscellaneous
