Towards Real-Time and Affordable Strain-Level Metagenomics-Based Foodborne Outbreak Investigations Using Oxford Nanopore Sequencing Technologies
- PMID: 34803953
- PMCID: PMC8602914
- DOI: 10.3389/fmicb.2021.738284
Towards Real-Time and Affordable Strain-Level Metagenomics-Based Foodborne Outbreak Investigations Using Oxford Nanopore Sequencing Technologies
Abstract
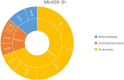
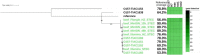
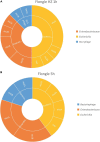
The current routine laboratory practices to investigate food samples in case of foodborne outbreaks still rely on attempts to isolate the pathogen in order to characterize it. We present in this study a proof of concept using Shiga toxin-producing Escherichia coli spiked food samples for a strain-level metagenomics foodborne outbreak investigation method using the MinION and Flongle flow cells from Oxford Nanopore Technologies, and we compared this to Illumina short-read-based metagenomics. After 12 h of MinION sequencing, strain-level characterization could be achieved, linking the food containing a pathogen to the related human isolate of the affected patient, by means of a single-nucleotide polymorphism (SNP)-based phylogeny. The inferred strain harbored the same virulence genes as the spiked isolate and could be serotyped. This was achieved by applying a bioinformatics method on the long reads using reference-based classification. The same result could be obtained after 24-h sequencing on the more recent lower output Flongle flow cell, on an extract treated with eukaryotic host DNA removal. Moreover, an alternative approach based on in silico DNA walking allowed to obtain rapid confirmation of the presence of a putative pathogen in the food sample. The DNA fragment harboring characteristic virulence genes could be matched to the E. coli genus after sequencing only 1 h with the MinION, 1 h with the Flongle if using a host DNA removal extraction, or 5 h with the Flongle with a classical DNA extraction. This paves the way towards the use of metagenomics as a rapid, simple, one-step method for foodborne pathogen detection and for fast outbreak investigation that can be implemented in routine laboratories on samples prepared with the current standard practices.
Keywords: Flongle; SNP analysis; STEC; food surveillance; metagenomics; nanopore; outbreak; strain-level.
Copyright © 2021 Buytaers, Saltykova, Denayer, Verhaegen, Vanneste, Roosens, Piérard, Marchal and De Keersmaecker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bogaerts B., Winand R., Fu Q., Van Braekel J., Ceyssens P. J., Mattheus W., et al. (2019). Validation of a bioinformatics workflow for routine analysis of whole-genome sequencing data and related challenges for pathogen typing in a European National Reference Center: Neisseria meningitidis as a proof-of-concept. Front. Microbiol. 10:362. 10.3389/fmicb.2019.00362 - DOI - PMC - PubMed
-
- Braeye T., Denayer S., De Rauw K., Forier A., Verluyten J., Fourie L., et al. (2014). Lessons learned from a textbook outbreak: EHEC-O157:H7 infections associated with the consumption of raw meat products, June 2012, Limburg, Belgium. Arch. Public Health 72:44. 10.1186/2049-3258-72-44 - DOI - PMC - PubMed
-
- Buytaers F. E., Saltykova A., Mattheus W., Verhaegen B., Roosens N. H. C., Vanneste K., et al. (2021b). Application of a strain-level shotgun metagenomics approach on food samples: resolution of the source of a Salmonella food-borne outbreak. Microb. Genom. 7:000547. 10.1099/mgen.0.000547 - DOI - PMC - PubMed
-
- Buytaers F. E., Fraiture M.-A., Berbers B., Vandermassen E., Hoffman S., Papazova N., et al. (2021a). A shotgun metagenomics approach to detect and characterize unauthorized genetically modified microorganisms in microbial fermentation products. Food Chem. Mol. Sci. 2:100023. 10.1016/j.fochms.2021.100023 - DOI - PMC - PubMed
-
- Buytaers F. E., Saltykova A., Denayer S., Verhaegen B., Vanneste K., Roosens N. H. C., et al. (2020). A practical method to implement strain-level metagenomics-based foodborne outbreak investigation and source tracking in routine. Microorganisms 8:1191. 10.3390/microorganisms8081191 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

