Small RNA Profiling in Mycobacterium Provides Insights Into Stress Adaptability
- PMID: 34803973
- PMCID: PMC8600241
- DOI: 10.3389/fmicb.2021.752537
Small RNA Profiling in Mycobacterium Provides Insights Into Stress Adaptability
Abstract
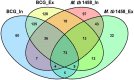
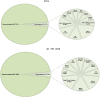
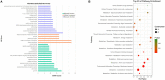
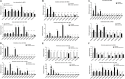
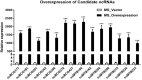
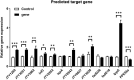
Mycobacteria encounter a number of environmental changes during infection and respond using different mechanisms. Small RNA (sRNA) is a post-transcriptionally regulatory system for gene functions and has been investigated in many other bacteria. This study used Mycobacterium tuberculosis and Mycobacterium bovis Bacillus Calmette-Guérin (BCG) infection models and sequenced whole bacterial RNAs before and after host cell infection. A comparison of differentially expressed sRNAs using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) and target prediction was carried out. Six pathogenically relevant stress conditions, growth rate, and morphology were used to screen and identify sRNAs. From these data, a subset of sRNAs was differentially expressed in multiple infection groups and stress conditions. Many were found associated with lipid metabolism. Among them, ncBCG427 was significantly downregulated when BCG entered into macrophages and was associated with increased biofilm formation. The reduction of virulence possibility depends on regulating lipid metabolism.
Keywords: Mycobacterium bovis; Mycobacterium bovis BCG; mycobacteria; sRNA; stress.
Copyright © 2021 Chen, Zhai, Zhang, Liu, Zhu, Su, Bermudez, Chen and Guo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources

