FlowKit: A Python Toolkit for Integrated Manual and Automated Cytometry Analysis Workflows
- PMID: 34804056
- PMCID: PMC8602902
- DOI: 10.3389/fimmu.2021.768541
FlowKit: A Python Toolkit for Integrated Manual and Automated Cytometry Analysis Workflows
Abstract
An important challenge for primary or secondary analysis of cytometry data is how to facilitate productive collaboration between domain and quantitative experts. Domain experts in cytometry laboratories and core facilities increasingly recognize the need for automated workflows in the face of increasing data complexity, but by and large, still conduct all analysis using traditional applications, predominantly FlowJo. To a large extent, this cuts domain experts off from the rapidly growing library of Single Cell Data Science algorithms available, curtailing the potential contributions of these experts to the validation and interpretation of results. To address this challenge, we developed FlowKit, a Gating-ML 2.0-compliant Python package that can read and write FCS files and FlowJo workspaces. We present examples of the use of FlowKit for constructing reporting and analysis workflows, including round-tripping results to and from FlowJo for joint analysis by both domain and quantitative experts.
Keywords: FlowJo; GatingML; flow cytometry; python (programming language); single cell data science; software; systems immunology.
Copyright © 2021 White, Quinn, Enzor, Staats, Mosier, Almarode, Denny, Weinhold, Ferrari and Chan.
Conflict of interest statement
Authors JQ and JA were employed by BD Biosciences - FlowJo. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

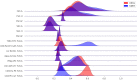
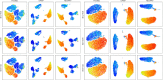


References
-
- Staats JS, Enzor JH, Sanchez AM, Rountree W, Chan C, Jaimes M, Chan RC, et al. . Toward Development of a Comprehensive External Quality Assurance Program for Polyfunctional Intracellular Cytokine Staining Assays. J Immunol Methods (2014) 409:44–53. doi: 10.1016/j.jim.2014.05.021 - DOI - PMC - PubMed
-
- Finak G, Jiang M. Flowworkspace: Infrastructure for Representing and Interacting With Gated and Ungated Cytometry Data Sets. In: R Package Version 4.4.0 (2021). doi: 10.18129/B9.bioc.flowWorkspace - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

