Database-Guided Analysis for Immunophenotypic Diagnosis and Follow-Up of Acute Myeloid Leukemia With Recurrent Genetic Abnormalities
- PMID: 34804933
- PMCID: PMC8602100
- DOI: 10.3389/fonc.2021.746951
Database-Guided Analysis for Immunophenotypic Diagnosis and Follow-Up of Acute Myeloid Leukemia With Recurrent Genetic Abnormalities
Abstract
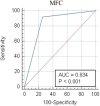
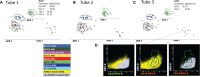
Acute myeloid leukemias (AMLs) are hematologic malignancies with varied molecular and immunophenotypic profiles, making them difficult to diagnose and classify. High-dimensional analysis algorithms might increase the utility of multicolor flow cytometry for AML diagnosis and follow-up. The objective of the present study was to assess whether a Compass database-guided analysis can be used to achieve rapid and accurate diagnoses. We conducted this study to determine whether this method could be employed to pilote the genetic and molecular tests and to objectively identify different-from-normal (DfN) patterns to improve measurable residual disease follow-up in AML. Three Compass databases were built using Infinicyt 2.0 software, including normal myeloid-committed hematopoietic precursors (n = 20) and AML blasts harboring the most frequent recurrent genetic abnormalities (n = 50). The diagnostic accuracy of the Compass database-guided analysis was evaluated in a prospective validation study (125 suspected AML patients). This method excluded AML associated with the following genetic abnormalities: t(8;21), t(15;17), inv(16), and KMT2A translocation, with 92% sensitivity [95% confidence interval (CI): 78.6%-98.3%] and a 98.5% negative predictive value (95% CI: 90.6%-99.8%). Our data showed that the Compass database-guided analysis could identify phenotypic differences between AML groups, representing a useful tool for the identification of DfN patterns.
Keywords: Compass database-guided analysis; acute myeloid leukemia with recurrent genetic abnormalities; different-from-normal (DfN) approach; measurable (minimal) residual disease; multicolor flow cytometry.
Copyright © 2021 Aanei, Veyrat-Masson, Selicean, Marian, Rigollet, Trifa, Tomuleasa, Serban, Cherry, Flandrin-Gresta, Tardy, Guyotat and Campos Catafal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Advanced Flow Cytometry Analysis Algorithms for Optimizing the Detection of "Different From Normal" Immunophenotypes in Acute Myeloid Blasts.Front Cell Dev Biol. 2021 Sep 28;9:735518. doi: 10.3389/fcell.2021.735518. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34650981 Free PMC article.
-
The predictive value of morphological findings in early diagnosis of acute myeloid leukemia with recurrent cytogenetic abnormalities.Leuk Res. 2018 Dec;75:23-28. doi: 10.1016/j.leukres.2018.10.017. Epub 2018 Nov 2. Leuk Res. 2018. PMID: 30445236
-
Flow cytometric analysis of acute leukemias. Diagnostic utility and critical analysis of data.Arch Pathol Lab Med. 2003 Jan;127(1):42-8. doi: 10.5858/2003-127-42-FCAOA. Arch Pathol Lab Med. 2003. PMID: 12521365
-
Acute Myeloid Leukemia: Diagnosis and Evaluation by Flow Cytometry.Cancers (Basel). 2024 Nov 17;16(22):3855. doi: 10.3390/cancers16223855. Cancers (Basel). 2024. PMID: 39594810 Free PMC article. Review.
-
Acute Myeloid Leukemia With Recurrent Cytogenetic Abnormalities.Am J Clin Pathol. 2015 Jul;144(1):6-18. doi: 10.1309/AJCPI9C8UILYQTNS. Am J Clin Pathol. 2015. PMID: 26071458 Review.
Cited by
-
Combined flow cytometry natural killer immunophenotyping and KIR/HLA-C genotyping reveal remarkable differences in acute myeloid leukemia patients, but suggest an overall impairment of the natural killer response.Front Med (Lausanne). 2023 Mar 7;10:1148748. doi: 10.3389/fmed.2023.1148748. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36960339 Free PMC article.
-
Identification of Leukemia-Associated Immunophenotypes by Databaseguided Flow Cytometry Provides a Highly Sensitive and Reproducible Strategy for the Study of Measurable Residual Disease in Acute Myeloblastic Leukemia.Cancers (Basel). 2022 Aug 19;14(16):4010. doi: 10.3390/cancers14164010. Cancers (Basel). 2022. PMID: 36011002 Free PMC article.
-
miR-379-5p Inhibited the Proliferation of Acute Myeloid Leukemia Cells Through Negative Regulation of YBX1.Turk J Haematol. 2025 May 22;42(2):92-99. doi: 10.4274/tjh.galenos.2025.2024.0424. Epub 2025 Feb 24. Turk J Haematol. 2025. PMID: 39992162 Free PMC article.
References
-
- van Dongen J, Lhermitte L, Böttcher S, Almeida J, van der Velden VHJ, Flores-Montero J, et al. . EuroFlow Antibody Panels for Standardised N-Dimensional Flow Cytometric Immunophenotyping of Normal, Reactive and Malignant Leukocytes. Leukemia (2012) 26:1908–75. doi: 10.1038/leu.2012.120 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

