A Comparative Proteomic Analysis of Extracellular Vesicles Associated With Lipotoxicity
- PMID: 34805145
- PMCID: PMC8600144
- DOI: 10.3389/fcell.2021.735001
A Comparative Proteomic Analysis of Extracellular Vesicles Associated With Lipotoxicity
Abstract
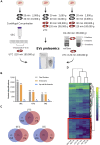
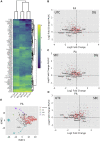
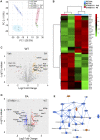
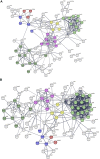
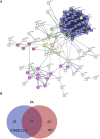
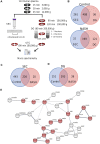
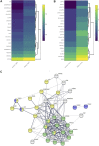
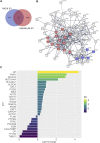
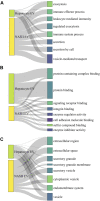
Extracellular vesicles (EVs) are emerging mediators of intercellular communication in nonalcoholic steatohepatitis (NASH). Palmitate, a lipotoxic saturated fatty acid, activates hepatocellular endoplasmic reticulum stress, which has been demonstrated to be important in NASH pathogenesis, including in the release of EVs. We have previously demonstrated that the release of palmitate-stimulated EVs is dependent on the de novo synthesis of ceramide, which is trafficked by the ceramide transport protein, STARD11. The trafficking of ceramide is a critical step in the release of lipotoxic EVs, as cells deficient in STARD11 do not release palmitate-stimulated EVs. Here, we examined the hypothesis that protein cargoes are trafficked to lipotoxic EVs in a ceramide-dependent manner. We performed quantitative proteomic analysis of palmitate-stimulated EVs in control and STARD11 knockout hepatocyte cell lines. Proteomics was performed on EVs isolated by size exclusion chromatography, ultracentrifugation, and density gradient separation, and EV proteins were measured by mass spectrometry. We also performed human EV proteomics from a control and a NASH plasma sample, for comparative analyses with hepatocyte-derived lipotoxic EVs. Size exclusion chromatography yielded most unique EV proteins. Ceramide-dependent lipotoxic EVs contain damage-associated molecular patterns and adhesion molecules. Haptoglobin, vascular non-inflammatory molecule-1, and insulin-like growth factor-binding protein complex acid labile subunit were commonly detected in NASH and hepatocyte-derived ceramide-dependent EVs. Lipotoxic EV proteomics provides novel candidate proteins to investigate in NASH pathogenesis and as diagnostic biomarkers for hepatocyte-derived EVs in NASH patients.
Keywords: DAMP; StAR-related lipid transfer domain 11; exosome; hepatocyte; microvesicle.
Copyright © 2021 Nakao, Fukushima, Mauer, Liao, Ferris, Dasgupta, Heppelmann, Vanderboom, Saraswat, Pandey, Nair, Allen, Nakao and Malhi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Aschrafi A., Franzen R., Shabahang S., Fabbro D., Pfeilschifter J., Huwiler A. (2003). Ceramide Induces Translocation of Protein Kinase C-α to the Golgi Compartment of Human Embryonic Kidney Cells by Interacting with the C2 Domain. Biochim. Biophys. Acta (Bba) - Mol. Cel Biol. Lipids 1634, 30–39. 10.1016/j.bbalip.2003.08.004 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

