Comprehensive Genetic Analysis Reveals Complexity of Monogenic Urinary Stone Disease
- PMID: 34805638
- PMCID: PMC8589729
- DOI: 10.1016/j.ekir.2021.08.033
Comprehensive Genetic Analysis Reveals Complexity of Monogenic Urinary Stone Disease
Abstract
Introduction: Because of phenotypic overlap between monogenic urinary stone diseases (USD), gene-specific analyses can result in missed diagnoses. We used targeted next generation sequencing (tNGS), including known and candidate monogenic USD genes, to analyze suspected primary hyperoxaluria (PH) or Dent disease (DD) patients genetically unresolved (negative; N) after Sanger analysis of the known genes. Cohorts consisted of 285 PH (PHN) and 59 DD (DDN) families.
Methods: Variants were assessed using disease-specific and population databases plus variant assessment tools and categorized using the American College of Medical Genetics (ACMG) guidelines. Prior Sanger analysis identified 47 novel PH or DD gene pathogenic variants.
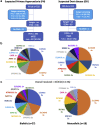
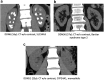
Results: Screening by tNGS revealed pathogenic variants in 14 known monogenic USD genes, accounting for 45 families (13.1%), 27 biallelic and 18 monoallelic, including 1 family with a copy number variant (CNV). Recurrent genes included the following: SLC34A3 (n = 13), CLDN16 (n = 8), CYP24A1 (n = 4), SLC34A1 (n = 3), SLC4A1 (n = 3), APRT (n = 2), CLDN19 (n = 2), HNF4A1 (n = 2), and KCNJ1 (n = 2), whereas ATP6V1B1, CASR, and SLC12A1 and missed CNVs in the PH genes AGXT and GRHPR accounted for 1 pedigree each. Of the 48 defined pathogenic variants, 27.1% were truncating and 39.6% were novel. Most patients were diagnosed before 18 years of age (76.1%), and 70.3% of biallelic patients were homozygous, mainly from consanguineous families.
Conclusion: Overall, in patients suspected of DD or PH, 23.9% and 7.3% of cases, respectively, were caused by pathogenic variants in other genes. This study shows the value of a tNGS screening approach to increase the diagnosis of monogenic USD, which can optimize therapies and facilitate enrollment in clinical trials.
Keywords: Dent disease; kidney stones; molecular genetics; monogenic; primary hyperoxaluria.
© 2021 International Society of Nephrology. Published by Elsevier Inc.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

