Geographic Patterns of Carbapenem-Resistant Pseudomonas aeruginosa in the Asia-Pacific Region: Results from the Antimicrobial Testing Leadership and Surveillance (ATLAS) Program, 2015-2019
- PMID: 34807753
- PMCID: PMC8846487
- DOI: 10.1128/AAC.02000-21
Geographic Patterns of Carbapenem-Resistant Pseudomonas aeruginosa in the Asia-Pacific Region: Results from the Antimicrobial Testing Leadership and Surveillance (ATLAS) Program, 2015-2019
Abstract
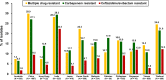
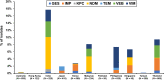
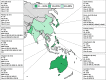
Pseudomonas aeruginosa is a common pathogen that is associated with multidrug-resistant (MDR) and carbapenem-resistant (CR) phenotypes; therefore, we investigated its resistance patterns and mechanisms by using data from the Antimicrobial Testing Leadership and Surveillance (ATLAS) program in the Asia-Pacific region from 2015 to 2019. MICs were determined using the broth microdilution method. Genes encoding major extended-spectrum β-lactamases and carbapenemases were investigated by multiplex PCR assays. Susceptibility was interpreted using the Clinical and Laboratory Standards Institute (CLSI) breakpoints. A total of 6,349 P. aeruginosa isolates were collected in the ATLAS program between 2015 and 2019 from 14 countries. According to the CLSI definitions, the numbers (and rates) of CR and MDR P. aeruginosa isolates were 1,198 (18.9%) and 1,303 (20.5%), respectively. For 747 of the CR P. aeruginosa strains that were available for gene screening, 253 β-lactamase genes were detected in 245 (32.8%) isolates. The most common gene was blaVIM (29.0%, 71/245), followed by blaNDM (24.9%, 61/245) and blaVEB (20.8%, 51/245). The resistance patterns and associated genes varied significantly between the countries in the Asia-Pacific region. India had the highest rates of carbapenem resistance (29.3%, 154/525) and gene detection (17.7%, 93/525). Compared to those harboring either class A or B β-lactamase genes, the CR P. aeruginosa isolates without detected β-lactamase genes had lower MICs for most of the antimicrobial agents, including ceftazidime-avibactam and ceftolozane-tazobactam. In conclusion, MDR and CR P. aeruginosa infections pose a major threat, particularly those with detected carbapenemase genes. Continuous surveillance is important for improving antimicrobial stewardship and antibiotic prescriptions.
Keywords: carbapenem resistant; carbapenemase; ceftazidime-avibactam; ceftolozane-tazobactam; multidrug resistant.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Elabbadi A, Pont S, Verdet C, Plésiat P, Cretin F, Voiriot G, Fartoukh M, Djibré M. 2020. An unusual community-acquired invasive and multi systemic infection due to ExoU-harboring Pseudomonas aeruginosa strain: clinical disease and microbiological characteristics. J Microbiol Immunol Infect 53:647–651. 10.1016/j.jmii.2019.06.008. - DOI - PubMed
-
- Viasus D, Puerta-Alcalde P, Cardozo C, Suárez-Lledó M, Rodríguez-Núñez O, Morata L, Fehér C, Marco F, Chumbita M, Moreno-García E, Fernández-Avilés F, Gutiérrez-Garcia G, Martínez JA, Mensa J, Rovira M, Esteve J, Soriano A, Garcia-Vidal C. 2020. Predictors of multidrug-resistant Pseudomonas aeruginosa in neutropenic patients with bloodstream infection. Clin Microbiol Infect 26:345–350. 10.1016/j.cmi.2019.07.002. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

