Origin and evolutionary landscape of Nr2f transcription factors across Metazoa
- PMID: 34807940
- PMCID: PMC8608329
- DOI: 10.1371/journal.pone.0254282
Origin and evolutionary landscape of Nr2f transcription factors across Metazoa
Abstract
Background: Nuclear Receptor Subfamily 2 Group F (Nr2f) orphan nuclear hormone transcription factors (TFs) are fundamental regulators of many developmental processes in invertebrates and vertebrates. Despite the importance of these TFs throughout metazoan development, previous work has not clearly outlined their evolutionary history.
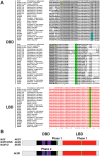
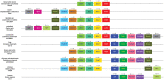
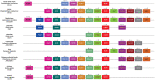
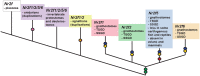
Results: We integrated molecular phylogeny with comparisons of intron/exon structure, domain architecture, and syntenic conservation to define critical evolutionary events that distinguish the Nr2f gene family in Metazoa. Our data indicate that a single ancestral eumetazoan Nr2f gene predated six main Bilateria subfamilies, which include single Nr2f homologs, here referred to as Nr2f1/2/5/6, that are present in invertebrate protostomes and deuterostomes, Nr2f1/2 homologs in agnathans, and Nr2f1, Nr2f2, Nr2f5, and Nr2f6 orthologs that are found in gnathostomes. Four cnidarian Nr2f1/2/5/6 and three agnathan Nr2f1/2 members are each due to independent expansions, while the vertebrate Nr2f1/Nr2f2 and Nr2f5/Nr2f6 members each form paralogous groups that arose from the established series of whole-genome duplications (WGDs). Nr2f6 members are the most divergent Nr2f subfamily in gnathostomes. Interestingly, in contrast to the other gnathostome Nr2f subfamilies, Nr2f5 has been independently lost in numerous vertebrate lineages. Furthermore, our analysis shows there are differential expansions and losses of Nr2f genes in teleosts following their additional rounds of WGDs.
Conclusion: Overall, our analysis of Nr2f gene evolution helps to reveal the origins and previously unrecognized relationships of this ancient TF family, which may allow for greater insights into the conservation of Nr2f functions that shape Metazoan body plans.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
-
- Pastorcic M, Wang H, Elbrecht A, Tsai SY, Tsai MJ, O’Malley BW. Control of transcription initiation in vitro requires binding of a transcription factor to the distal promoter of the ovalbumin gene. Molecular and Cellular Biology. 1986. pp. 2784–2791. doi: 10.1128/mcb.6.8.2784-2791.1986 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

