Novel Filoviruses, Hantavirus, and Rhabdovirus in Freshwater Fish, Switzerland, 2017
- PMID: 34808081
- PMCID: PMC8632185
- DOI: 10.3201/eid2712.210491
Novel Filoviruses, Hantavirus, and Rhabdovirus in Freshwater Fish, Switzerland, 2017
Abstract
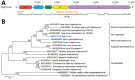
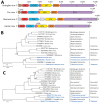
European perch (Perca fluviatilis) are increasingly farmed as a human food source. Viral infections of European perch remain largely unexplored, thereby putting farm populations at incalculable risk for devastating fish epizootics and presenting a potential hazard to consumers. To address these concerns, we applied metatranscriptomics to identify disease-associated viruses in European perch farmed in Switzerland. Unexpectedly, in clinically diseased fish we detected novel freshwater fish filoviruses, a novel freshwater fish hantavirus, and a previously unknown rhabdovirus. Hantavirus titers were high, and we demonstrated virus in macrophages and gill endothelial cells by using in situ hybridization. Rhabdovirus titers in organ samples were low, but virus could be isolated on cell culture. Our data add to the hypothesis that filoviruses, hantaviruses, and rhabdoviruses are globally distributed common fish commensals, pathogens, or both. Our findings shed new light on negative-sense RNA virus diversity and evolution.
Keywords: European perch; Switzerland; filovirus; freshwater fish; hantavirus; rhabdovirus; viruses.
Figures


Similar articles
-
Genetic diversity and associated pathology of rhabdovirus infections in farmed and wild perch Perca fluviatilis in Ireland.Dis Aquat Organ. 2014 Dec 2;112(2):121-30. doi: 10.3354/dao02801. Dis Aquat Organ. 2014. PMID: 25449323
-
First isolation of a rhabdovirus from perch Perca fluviatilis in Switzerland.Dis Aquat Organ. 2015 Oct 16;116(2):93-101. doi: 10.3354/dao02908. Dis Aquat Organ. 2015. PMID: 26480912
-
Genetic diversity of perch rhabdoviruses isolates based on the nucleoprotein and glycoprotein genes.Arch Virol. 2011 Dec;156(12):2133-44. doi: 10.1007/s00705-011-1103-z. Epub 2011 Sep 17. Arch Virol. 2011. PMID: 21927897
-
Fish rhabdoviruses: molecular epidemiology and evolution.Curr Top Microbiol Immunol. 2005;292:81-117. doi: 10.1007/3-540-27485-5_5. Curr Top Microbiol Immunol. 2005. PMID: 15981469 Review.
-
Immunity to fish rhabdoviruses.Viruses. 2012 Jan;4(1):140-66. doi: 10.3390/v4010140. Epub 2012 Jan 18. Viruses. 2012. PMID: 22355456 Free PMC article. Review.
Cited by
-
Five Species of Wild Freshwater Sport Fish in Wisconsin, USA, Reveal Highly Diverse Viromes.Pathogens. 2024 Feb 7;13(2):150. doi: 10.3390/pathogens13020150. Pathogens. 2024. PMID: 38392888 Free PMC article.
-
Novel filoviruses: indication of a global threat or cause to reassess our risk perception?Npj Viruses. 2024 Aug 21;2(1):38. doi: 10.1038/s44298-024-00050-4. Npj Viruses. 2024. PMID: 40295872 Free PMC article. Review.
-
Genomic transfers help to decipher the ancient evolution of filoviruses and interactions with vertebrate hosts.PLoS Pathog. 2024 Sep 3;20(9):e1011864. doi: 10.1371/journal.ppat.1011864. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39226335 Free PMC article.
-
Attempted Transmission of Marburg Virus by Bat-Associated Fleas Thaumapsylla breviceps breviceps (Ischnopsyllidae: Thaumapsyllinae) to the Egyptian Rousette Bat (Rousettus aegyptiacus).Viruses. 2024 Jul 25;16(8):1197. doi: 10.3390/v16081197. Viruses. 2024. PMID: 39205171 Free PMC article.
-
A novel group of negative-sense RNA viruses associated with epizootics in managed and free-ranging freshwater turtles in Florida, USA.PLoS Pathog. 2022 Mar 11;18(3):e1010258. doi: 10.1371/journal.ppat.1010258. eCollection 2022 Mar. PLoS Pathog. 2022. PMID: 35275967 Free PMC article.
References
-
- Food and Agriculture Organization of the United Nations. The state of world fisheries and aquaculture 2018. Meeting the sustainable development goals [cited 2021 Jul 10]. https://www.fao.org/3/i9540en/I9540EN.pdf
-
- Martins CIM, Eding EH, Verdegem MCJ, Heinsbroek LTN, Schneider O, Blancheton JP, et al. New developments in recirculating aquaculture systems in Europe: a perspective on environmental sustainability. Aquacult Eng. 2010;43:83–93. 10.1016/j.aquaeng.2010.09.002 - DOI
-
- Policar T, Schaefer FJ, Panana E, Meyer S, Teerlinck S, Toner D, et al. Recent progress in European percid fish culture production technology—tackling bottlenecks. Aquacult Int. 2019;27:1151–74. 10.1007/s10499-019-00433-y - DOI
-
- Morgan DL, Gill HS, Maddern MG, Beatty SJ. Distribution and impacts of introduced freshwater fishes in Western Australia. N J Mar Freshwater Res. 2004;38:511–23. 10.1080/00288330.2004.9517257 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

