Influenza A M2 recruits M1 to the plasma membrane: A fluorescence fluctuation microscopy study
- PMID: 34808098
- PMCID: PMC8715234
- DOI: 10.1016/j.bpj.2021.11.023
Influenza A M2 recruits M1 to the plasma membrane: A fluorescence fluctuation microscopy study
Abstract
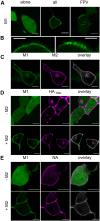
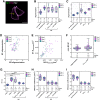
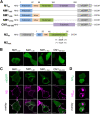
Influenza A virus (IAV) is a respiratory pathogen that causes seasonal epidemics with significant mortality. One of the most abundant proteins in IAV particles is the matrix protein 1 (M1), which is essential for the virus structural stability. M1 organizes virion assembly and budding at the plasma membrane (PM), where it interacts with other viral components. The recruitment of M1 to the PM as well as its interaction with the other viral envelope proteins (hemagglutinin [HA], neuraminidase, matrix protein 2 [M2]) is controversially discussed in previous studies. Therefore, we used fluorescence fluctuation microscopy techniques (i.e., scanning fluorescence cross-correlation spectroscopy and number and brightness) to quantify the oligomeric state of M1 and its interactions with other viral proteins in co-transfected as well as infected cells. Our results indicate that M1 is recruited to the PM by M2, as a consequence of the strong interaction between the two proteins. In contrast, only a weak interaction between M1 and HA was observed. M1-HA interaction occurred only in the event that M1 was already bound to the PM. We therefore conclude that M2 initiates the assembly of IAV by recruiting M1 to the PM, possibly allowing its further interaction with other viral proteins.
Copyright © 2021 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Davlin S., Blanton L., et al. Brammer L. Influenza activity - United States, 2015-16 season and composition of the 2016-17 influenza vaccine. Morb. Mortal. Wkly. Rep. 2016;65:567–575. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

