Vertical Inheritance Facilitates Interspecies Diversification in Biosynthetic Gene Clusters and Specialized Metabolites
- PMID: 34809466
- PMCID: PMC8609351
- DOI: 10.1128/mBio.02700-21
Vertical Inheritance Facilitates Interspecies Diversification in Biosynthetic Gene Clusters and Specialized Metabolites
Abstract
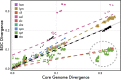
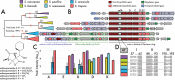
While specialized metabolites are thought to mediate ecological interactions, the evolutionary processes driving chemical diversification, particularly among closely related lineages, remain poorly understood. Here, we examine the evolutionary dynamics governing the distribution of natural product biosynthetic gene clusters (BGCs) among 118 strains representing all nine currently named species of the marine actinobacterial genus Salinispora. While much attention has been given to the role of horizontal gene transfer (HGT) in structuring BGC distributions, we find that vertical descent facilitates interspecies BGC diversification over evolutionary timescales. Moreover, we identified a distinct phylogenetic signal among Salinispora species at both the BGC and metabolite level, indicating that specialized metabolism represents a conserved phylogenetic trait. Using a combination of genomic analyses and liquid chromatography-high-resolution tandem mass spectrometry (LC-MS/MS) targeting nine experimentally characterized BGCs and their small molecule products, we identified gene gain/loss events, constrained interspecies recombination, and other evolutionary processes associated with vertical inheritance as major contributors to BGC diversification. These evolutionary dynamics had direct consequences for the compounds produced, as exemplified by species-level differences in salinosporamide production. Together, our results support the concept that specialized metabolites, and their cognate BGCs, can represent phylogenetically conserved functional traits with chemical diversification proceeding in species-specific patterns over evolutionary time frames. IMPORTANCE Microbial natural products are traditionally exploited for their pharmaceutical potential, yet our understanding of the evolutionary processes driving BGC evolution and compound diversification remain poorly developed. While HGT is recognized as an integral driver of BGC distributions, we find that the effects of vertical inheritance on BGC diversification had direct implications for species-level specialized metabolite production. As such, understanding the degree of genetic variation that corresponds to species delineations can enhance natural product discovery efforts. Resolving the evolutionary relationships between closely related strains and specialized metabolism can also facilitate our understanding of the ecological roles of small molecules in structuring the environmental distribution of microbes.
Keywords: Salinispora; evolution; evolutionary biology; homologous recombination; microbial ecology; salinosporamide.
Conflict of interest statement
We declare that there is no conflict of interest regarding the publication of this article.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
