Stabilization of UCA1 by N6-methyladenosine RNA methylation modification promotes colorectal cancer progression
- PMID: 34809621
- PMCID: PMC8609784
- DOI: 10.1186/s12935-021-02288-x
Stabilization of UCA1 by N6-methyladenosine RNA methylation modification promotes colorectal cancer progression
Abstract
Background: UCA1 is frequently upregulated in a variety of cancers, including CRC, and it can play an oncogenic role by various mechanisms. However, how UCA1 is regulated in cancer is largely unknown. In this study, we aimed to determine whether RNA methylation at N6-methyladenosine (m6A) can impact UCA1 expression in colorectal cancer (CRC).
Methods: qRT-PCR was performed to detect the level of UCA1 and IGF2BP2 in CRC samples. CRISPR/Cas9 was employed to knockout (KO) UCA1, METTL3 and WTAP in DLD-1 and HCT-116 cells, while rescue experiments were carried out to re-express METTL3 and WTAP in KO cells. Immunoprecipitation using m6A antibody was performed to determine the m6A modification of UCA1. In vivo pulldown assays using S1m tagging combined with site-direct mutagenesis was carried out to confirm the recognition of m6A-modified UCA1 by IGF2BP2. Cell viability was measured by MTT and colony formation assays. The expression of UCA1 and IGF2BP2 in TCGA CRC database was obtained from GEPIA ( http://gepia.cancer-pku.cn ).
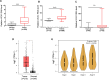
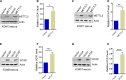
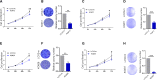
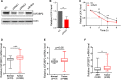
Results: Our results revealed that IGF2BP2 serves as a reader for m6A modified UCA1 and that adenosine at 1038 of UCA1 is critical to the recognition by IGF2BP2. Importantly, we showed that m6A writers, METTL3 and WTAP positively regulate UCA1 expression. Mechanically, IGF2BP2 increases the stability of m6A-modified UCA1. Clinically, IGF2BP2 is upregulated in CRC tissues compared with normal tissues.
Conclusion: These results suggest that m6A modification is an important factor contributing to upregulation of UCA1 in CRC tissues.
Keywords: CRC; IGF2BP2; UCA1; m6A modification.
© 2021. The Author(s).
Conflict of interest statement
The authors declared no competing interests in this study.
Figures




Similar articles
-
METTL3/IGF2BP2 axis affects the progression of colorectal cancer by regulating m6A modification of STAG3.Sci Rep. 2023 Oct 12;13(1):17292. doi: 10.1038/s41598-023-44379-x. Sci Rep. 2023. PMID: 37828232 Free PMC article.
-
The m6A Reader IGF2BP2 Promotes Oral Squamous Cell Carcinoma Progression by Maintaining UCA1 Stability.Recent Pat Anticancer Drug Discov. 2024 Aug 26. doi: 10.2174/0115748928293003240817180839. Online ahead of print. Recent Pat Anticancer Drug Discov. 2024. PMID: 39192648
-
RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner.J Exp Clin Cancer Res. 2022 Jan 3;41(1):6. doi: 10.1186/s13046-021-02212-1. J Exp Clin Cancer Res. 2022. PMID: 34980207 Free PMC article.
-
The potential role of RNA N6-methyladenosine in Cancer progression.Mol Cancer. 2020 May 12;19(1):88. doi: 10.1186/s12943-020-01204-7. Mol Cancer. 2020. PMID: 32398132 Free PMC article. Review.
-
The functions of N6-methyladenosine (m6A) RNA modifications in colorectal cancer.Med Oncol. 2022 Sep 29;39(12):235. doi: 10.1007/s12032-022-01827-4. Med Oncol. 2022. PMID: 36175777 Review.
Cited by
-
Role of WTAP in Cancer: From Mechanisms to the Therapeutic Potential.Biomolecules. 2022 Sep 2;12(9):1224. doi: 10.3390/biom12091224. Biomolecules. 2022. PMID: 36139062 Free PMC article. Review.
-
Non-coding RNAs as therapeutic targets in cancer and its clinical application.J Pharm Anal. 2024 Jul;14(7):100947. doi: 10.1016/j.jpha.2024.02.001. Epub 2024 Feb 8. J Pharm Anal. 2024. PMID: 39149142 Free PMC article. Review.
-
The Role of m6A Epigenetic Modification in the Treatment of Colorectal Cancer Immune Checkpoint Inhibitors.Front Immunol. 2022 Jan 6;12:802049. doi: 10.3389/fimmu.2021.802049. eCollection 2021. Front Immunol. 2022. PMID: 35069586 Free PMC article. Review.
-
YTHDC1 negatively regulates UBE3A to influence RAD51 ubiquitination and inhibit apoptosis in colorectal cancer cells.Sci Rep. 2025 Mar 14;15(1):8857. doi: 10.1038/s41598-025-92925-6. Sci Rep. 2025. PMID: 40087295 Free PMC article.
-
Focused ultrasound combined with miR-1208-equipped exosomes inhibits malignant progression of glioma.Br J Cancer. 2023 Oct;129(7):1083-1094. doi: 10.1038/s41416-023-02393-w. Epub 2023 Aug 14. Br J Cancer. 2023. PMID: 37580442 Free PMC article.
References
-
- Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. - PubMed
-
- Wu C, Li M, Meng H, Liu Y, Niu W, Zhou Y, Zhao R, Duan Y, Zeng Z, Li X, et al. Analysis of status and countermeasures of cancer incidence and mortality in China. Sci China Life Sci. 2019;62(5):640–647. - PubMed
-
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. - PubMed
-
- Siegel RL, Torre LA, Soerjomataram I, Hayes RB, Bray F, Weber TK, Jemal A. Global patterns and trends in colorectal cancer incidence in young adults. Gut. 2019;68(12):2179–2185. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

