Methodology for clinical genotyping of CYP2D6 and CYP2C19
- PMID: 34811360
- PMCID: PMC8608805
- DOI: 10.1038/s41398-021-01717-9
Methodology for clinical genotyping of CYP2D6 and CYP2C19
Erratum in
-
Correction: Methodology for clinical genotyping of CYP2D6 and CYP2C19.Transl Psychiatry. 2022 Mar 8;12(1):94. doi: 10.1038/s41398-022-01845-w. Transl Psychiatry. 2022. PMID: 35260547 Free PMC article. No abstract available.
Abstract
Many antidepressants, atomoxetine, and several antipsychotics are metabolized by the cytochrome P450 enzymes CYP2D6 and CYP2C19, and guidelines for prescribers based on genetic variants exist. Although some laboratories offer such testing, there is no consensus regarding validated methodology for clinical genotyping of CYP2D6 and CYP2C19. The aim of this paper was to cross-validate multiple technologies for genotyping CYP2D6 and CYP2C19 against each other, and to contribute to feasibility for clinical implementation by providing an enhanced range of assay options, customizable automated translation of data into haplotypes, and a workflow algorithm. AmpliChip CYP450 and some TaqMan single nucleotide variant (SNV) and copy number variant (CNV) data in the Genome-based therapeutic drugs for depression (GENDEP) study were used to select 95 samples (out of 853) to represent as broad a range of CYP2D6 and CYP2C19 genotypes as possible. These 95 included a larger range of CYP2D6 hybrid configurations than have previously been reported using inter-technology data. Genotyping techniques employed were: further TaqMan CNV and SNV assays, xTAGv3 Luminex CYP2D6 and CYP2C19, PharmacoScan, the Ion AmpliSeq Pharmacogenomics Panel, and, for samples with CYP2D6 hybrid configurations, long-range polymerase chain reactions (L-PCRs) with Sanger sequencing and Luminex. Agena MassARRAY was also used for CYP2C19. This study has led to the development of a broader range of TaqMan SNV assays, haplotype phasing methodology with TaqMan adaptable for other technologies, a multiplex genotyping method for efficient identification of some hybrid haplotypes, a customizable automated translation of SNV and CNV data into haplotypes, and a clinical workflow algorithm.
© 2021. The Author(s).
Conflict of interest statement
NH has participated in research supported by CSF project No. IP-09-2014-2979. DS has received grant/research support from Janssen and Lundbeck and has served as a consultant or on advisory boards for Janssen and Lundbeck. KJA is a member of the Clinical Pharmacogenetics Implementation Consortium and the Pharmacogene Variation Consortium, has received two research grants in the last two years from Janssen Inc., Canada (fellowship grants for trainees). All other authors have nothing to disclose.
Figures
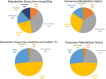

References
-
- Aitchison KJ, Jordan BD, Sharma T. The relevance of ethnic influences on pharmacogenetics to the treatment of psychosis. Drug Metab Drug Interact. 2000;16:15–38. - PubMed
-
- Lapetina DL, Yang EH, Henriques BC, Aitchison KJ. Pharmacogenomics and psychopharmacology. In: Haddad PM, Nutt DJ, editors. Seminars in clinical psychopharmacology. London: Cambridge University Press; 2020. pp. 153–204.
-
- Jukic MM, Haslemo T, Molden E, Ingelman-Sundberg M. Impact of CYP2C19 genotype on escitalopram exposure and therapeutic failure: a Retrospective Study based on 2,087 patients. Am J Psychiatry. 2018;175:463–70. - PubMed

