Dynamic changes in immune gene co-expression networks predict development of type 1 diabetes
- PMID: 34811390
- PMCID: PMC8609030
- DOI: 10.1038/s41598-021-01840-z
Dynamic changes in immune gene co-expression networks predict development of type 1 diabetes
Abstract
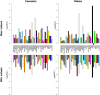
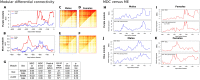
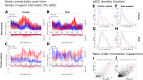
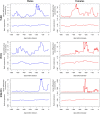
Significant progress has been made in elucidating genetic risk factors influencing Type 1 diabetes (T1D); however, features other than genetic variants that initiate and/or accelerate islet autoimmunity that lead to the development of clinical T1D remain largely unknown. We hypothesized that genetic and environmental risk factors can both contribute to T1D through dynamic alterations of molecular interactions in physiologic networks. To test this hypothesis, we utilized longitudinal blood transcriptomic profiles in The Environmental Determinants of Diabetes in the Young (TEDDY) study to generate gene co-expression networks. In network modules that contain immune response genes associated with T1D, we observed highly dynamic differences in module connectivity in the 600 days (~ 2 years) preceding clinical diagnosis of T1D. Our results suggest that gene co-expression is highly plastic and that connectivity differences in T1D-associated immune system genes influence the timing and development of clinical disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

