The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants
- PMID: 34812570
- PMCID: PMC8989497
- DOI: 10.1111/pbi.13751
The GhMYB36 transcription factor confers resistance to biotic and abiotic stress by enhancing PR1 gene expression in plants
Abstract
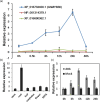
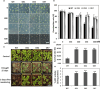
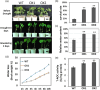
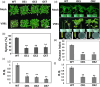
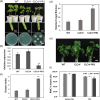
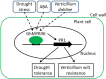
Drought and Verticillium wilt disease are two main factors that limit cotton production, which necessitates the identification of key molecular switch to simultaneously improve cotton resistance to Verticillium dahliae and tolerance to drought stress. R2R3-type MYB proteins could play such a role because of their conserved functions in plant development, growth, and metabolism regulation, however, till date a MYB gene conferring the desired resistance to both biotic and abiotic stresses has not been found in cotton. Here, we describe the identification of GhMYB36, a gene encoding a R2R3-type MYB protein in Gossypium hirsutum, which confers drought tolerance and Verticilium wilt resistance in both Arabidopsis and cotton. GhMYB36 was highly induced by PEG-simulated drought stress in G. hirsutum. GhMYB36-silenced cotton plants were more sensitive to both drought stress and Verticillium wilt. GhMYB36 overexpression in transgenic Arabidopsis and cotton plants gave rise to improved drought tolerance and Verticillium wilt resistance. Transient expression of fused GhMYB36-GFP in tobacco cells was able to localize GhMYB36 in the cell nucleus. In addition, RNA-seq analysis together with qRT-PCR validation in transgenic Arabidopsis overexpressing GhMYB36 revealed significantly enhanced PR1 expression. Luciferase interaction assays indicated that GhMYB36 are probably bound to the promoter of PR1 to activate its expression and the interaction, which was further verified by Yeast one hybrid assay. Taken together, our results suggest that GhMYB36 functions as a transcription factor that is involved in drought tolerance and Verticillium wilt resistance in Arabidopsis and cotton by enhancing PR1 expression.
Keywords: R2R3-type MYB proteins; drought tolerance; transcription factors; transgenic plants; transient expression.
© 2021 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
No conflicts of interest declared.
Figures



References
-
- Akbudak, M.A. , Yildiz, S. and Filiz, E. (2020) Pathogenesis related protein‐1 (PR‐1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics, 112, 4089–4099. - PubMed
-
- Chen, T. , Li, W. , Hu, X. , Guo, J. , Liu, A. and Zhang, B. (2015) A cotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol., 56, 917–929. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

