Identification of isolated or mixed strains from long reads: a challenge met on Streptococcus thermophilus using a MinION sequencer
- PMID: 34812718
- PMCID: PMC8743539
- DOI: 10.1099/mgen.0.000654
Identification of isolated or mixed strains from long reads: a challenge met on Streptococcus thermophilus using a MinION sequencer
Abstract
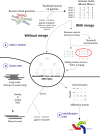
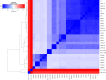
This study aimed to provide efficient recognition of bacterial strains on personal computers from MinION (Nanopore) long read data. Thanks to the fall in sequencing costs, the identification of bacteria can now proceed by whole genome sequencing. MinION is a fast, but highly error-prone sequencing device and it is a challenge to successfully identify the strain content of unknown simple or complex microbial samples. It is heavily constrained by memory management and fast access to the read and genome fragments. Our strategy involves three steps: indexing of known genomic sequences for a given or several bacterial species; a request process to assign a read to a strain by matching it to the closest reference genomes; and a final step looking for a minimum set of strains that best explains the observed reads. We have applied our method, called ORI, on 77 strains of Streptococcus thermophilus. We worked on several genomic distances and obtained a detailed classification of the strains, together with a criterion that allows merging of what we termed 'sibling' strains, only separated by a few mutations. Overall, isolated strains can be safely recognized from MinION data. For mixtures of several non-sibling strains, results depend on strain abundance.
Keywords: MinION; Streptococcus thermophilus; bacterial strain identification; bloom filters; long read; strain classification.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing.BMC Genomics. 2019 Jan 9;20(1):23. doi: 10.1186/s12864-018-5381-7. BMC Genomics. 2019. PMID: 30626323 Free PMC article.
-
The use of Oxford Nanopore native barcoding for complete genome assembly.Gigascience. 2017 Mar 1;6(3):1-6. doi: 10.1093/gigascience/gix001. Gigascience. 2017. PMID: 28327913 Free PMC article.
-
Genome assembly using Nanopore-guided long and error-free DNA reads.BMC Genomics. 2015 Apr 20;16(1):327. doi: 10.1186/s12864-015-1519-z. BMC Genomics. 2015. PMID: 25927464 Free PMC article.
-
Adaptation of Oxford Nanopore technology for hepatitis C whole genome sequencing and identification of within-host viral variants.BMC Genomics. 2021 Mar 2;22(1):148. doi: 10.1186/s12864-021-07460-1. BMC Genomics. 2021. PMID: 33653280 Free PMC article.
-
On-Site MinION Sequencing.Adv Exp Med Biol. 2019;1129:143-150. doi: 10.1007/978-981-13-6037-4_10. Adv Exp Med Biol. 2019. PMID: 30968366 Review.
Cited by
-
The genomic basis of the Streptococcus thermophilus health-promoting properties.BMC Genomics. 2022 Mar 16;23(1):210. doi: 10.1186/s12864-022-08459-y. BMC Genomics. 2022. PMID: 35291951 Free PMC article.
-
A step forward for Shiga toxin-producing Escherichia coli identification and characterization in raw milk using long-read metagenomics.Microb Genom. 2022 Nov;8(11):mgen000911. doi: 10.1099/mgen.0.000911. Microb Genom. 2022. PMID: 36748417 Free PMC article.
-
cgMSI: pathogen detection within species from nanopore metagenomic sequencing data.BMC Bioinformatics. 2023 Oct 12;24(1):387. doi: 10.1186/s12859-023-05512-9. BMC Bioinformatics. 2023. PMID: 37821827 Free PMC article.
-
Comparative genome analysis of 15 Streptococcus thermophilus strains isolated from Turkish traditional yogurt.Antonie Van Leeuwenhoek. 2025 Mar 28;118(4):64. doi: 10.1007/s10482-025-02070-3. Antonie Van Leeuwenhoek. 2025. PMID: 40153053
References
-
- Siezen RJ, Starrenburg MJC, Boekhorst J, Renckens B, Molenaar D, et al. Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl Environ Microbiol. 2008;74:424–436. doi: 10.1128/AEM.01850-07. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

