An NR2F1-specific agonist suppresses metastasis by inducing cancer cell dormancy
- PMID: 34812843
- PMCID: PMC8614154
- DOI: 10.1084/jem.20210836
An NR2F1-specific agonist suppresses metastasis by inducing cancer cell dormancy
Abstract
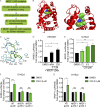
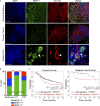
We describe the discovery of an agonist of the nuclear receptor NR2F1 that specifically activates dormancy programs in malignant cells. The agonist led to a self-regulated increase in NR2F1 mRNA and protein and downstream transcription of a novel dormancy program. This program led to growth arrest of an HNSCC PDX line, human cell lines, and patient-derived organoids in 3D cultures and in vivo. This effect was lost when NR2F1 was knocked out by CRISPR-Cas9. RNA sequencing revealed that agonist treatment induces transcriptional changes associated with inhibition of cell cycle progression and mTOR signaling, metastasis suppression, and induction of a neural crest lineage program. In mice, agonist treatment resulted in inhibition of lung HNSCC metastasis, even after cessation of the treatment, where disseminated tumor cells displayed an NR2F1hi/p27hi/Ki-67lo/p-S6lo phenotype and remained in a dormant single-cell state. Our work provides proof of principle supporting the use of NR2F1 agonists to induce dormancy as a therapeutic strategy to prevent metastasis.
© 2021 Khalil et al.
Conflict of interest statement
Disclosures: J.A. Aguirre-Ghiso reported grants from HiberCell LLC during the conduct of the study; grants from HiberCell LLC outside the submitted work; and is a HiberCell LLC co-founder, consultant, and scientific advisory board member. No other disclosures were reported.
Figures










References
-
- Adam, A.P., George A., Schewe D., Bragado P., Iglesias B.V., Ranganathan A.C., Kourtidis A., Conklin D.S., and Aguirre-Ghiso J.A.. 2009. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Res. 69:5664–5672. 10.1158/0008-5472.CAN-08-3820 - DOI - PMC - PubMed
-
- Aguirre-Ghiso, J.A., Ossowski L., and Rosenbaum S.K.. 2004. Green fluorescent protein tagging of extracellular signal-regulated kinase and p38 pathways reveals novel dynamics of pathway activation during primary and metastatic growth. Cancer Res. 64:7336–7345. 10.1158/0008-5472.CAN-04-0113 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA109182/CA/NCI NIH HHS/United States
- CA078207/CA/NCI NIH HHS/United States
- R01 CA218024/CA/NCI NIH HHS/United States
- R01 CA216248/CA/NCI NIH HHS/United States
- R01 CA109182/CA/NCI NIH HHS/United States
- T32 CA078207/CA/NCI NIH HHS/United States
- K22 CA201054/CA/NCI NIH HHS/United States
- S10OD018522/NH/NIH HHS/United States
- CCR17483357/KOMEN/Susan G. Komen/United States
- S10 OD018522/OD/NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- P30 CA196521/CA/NCI NIH HHS/United States
- CA201054/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

