Customization of a DADA2-based pipeline for fungal internal transcribed spacer 1 (ITS1) amplicon data sets
- PMID: 34813499
- PMCID: PMC8765055
- DOI: 10.1172/jci.insight.151663
Customization of a DADA2-based pipeline for fungal internal transcribed spacer 1 (ITS1) amplicon data sets
Abstract
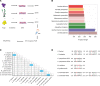
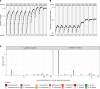
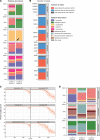
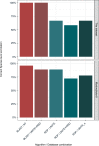
Identification and analysis of fungal communities commonly rely on internal transcribed spacer-based (ITS-based) amplicon sequencing. There is no gold standard used to infer and classify fungal constituents since methodologies have been adapted from analyses of bacterial communities. To achieve high-resolution inference of fungal constituents, we customized a DADA2-based pipeline using a mix of 11 medically relevant fungi. While DADA2 allowed the discrimination of ITS1 sequences differing by single nucleotides, quality filtering, sequencing bias, and database selection were identified as key variables determining the accuracy of sample inference. Due to species-specific differences in sequencing quality, default filtering settings removed most reads that originated from Aspergillus species, Saccharomyces cerevisiae, and Candida glabrata. By fine-tuning the quality filtering process, we achieved an improved representation of the fungal communities. By adapting a wobble nucleotide in the ITS1 forward primer region, we further increased the yield of S. cerevisiae and C. glabrata sequences. Finally, we showed that a BLAST-based algorithm based on the UNITE+INSD or the NCBI NT database achieved a higher reliability in species-level taxonomic annotation compared with the naive Bayesian classifier implemented in DADA2. These steps optimized a robust fungal ITS1 sequencing pipeline that, in most instances, enabled species-level assignment of community members.
Keywords: Fungal infections; Infectious disease; Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

