ACC1-expressing pathogenic T helper 2 cell populations facilitate lung and skin inflammation in mice
- PMID: 34813654
- PMCID: PMC8614157
- DOI: 10.1084/jem.20210639
ACC1-expressing pathogenic T helper 2 cell populations facilitate lung and skin inflammation in mice
Abstract
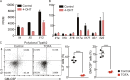
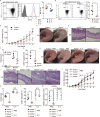
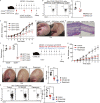
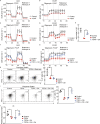
T cells possess distinguishing effector functions and drive inflammatory disorders. We have previously identified IL-5-producing Th2 cells as the pathogenic population predominantly involved in the pathology of allergic inflammation. However, the cell-intrinsic signaling pathways that control the pathogenic Th2 cell function are still unclear. We herein report the high expression of acetyl-CoA carboxylase 1 (ACC1) in the pathogenic CD4+ T cell population in the lung and skin. The genetic deletion of CD4+ T cell-intrinsic ACC1 dampened eosinophilic and basophilic inflammation in the lung and skin by constraining IL-5 or IL-3 production. Mechanistically, ACC1-dependent fatty acid biosynthesis induces the pathogenic cytokine production of CD4+ T cells via metabolic reprogramming and the availability of acetyl-CoA for epigenetic regulation. We thus identified a distinct phenotype of the pathogenic T cell population in the lung and skin, and ACC1 was shown to be an essential regulator controlling the pathogenic function of these populations to promote type 2 inflammation.
© 2021 Nakajima et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures










References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

