Multicenter assessment of shotgun metagenomics for pathogen detection
- PMID: 34814051
- PMCID: PMC8608867
- DOI: 10.1016/j.ebiom.2021.103649
Multicenter assessment of shotgun metagenomics for pathogen detection
Abstract
Background: Shotgun metagenomics has been used clinically for diagnosing infectious diseases. However, most technical assessments have been limited to individual sets of reference standards, experimental workflows, and laboratories.
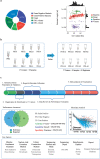
Methods: A reference panel and performance metrics were designed and used to examine the performance of shotgun metagenomics at 17 laboratories in a coordinated collaborative study. We comprehensively assessed the reliability, key performance determinants, reproducibility, and quantitative potential.
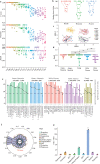
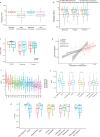
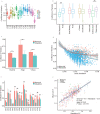
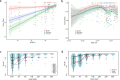
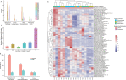
Findings: Assay performance varied significantly across sites and microbial classes, with a read depth of 20 millions as a generally cost-efficient assay setting. Results of mapped reads by shotgun metagenomics could indicate relative and intra-site (but not absolute or inter-site) microbial abundance.
Interpretation: Assay performance was significantly impacted by the microbial type, the host context, and read depth, which emphasizes the importance of these factors when designing reference reagents and benchmarking studies. Across sites, workflows and platforms, false positive reporting and considerable site/library effects were common challenges to the assay's accuracy and quantifiability. Our study also suggested that laboratory-developed shotgun metagenomics tests for pathogen detection should aim to detect microbes at 500 CFU/mL (or copies/mL) in a clinically relevant host context (10^5 human cells/mL) within a 24h turn-around time, and with an efficient read depth of 20M.
Funding: This work was supported by National Science and Technology Major Project of China (2018ZX10102001).
Keywords: Diagnostic Performance; Multicenter Assessment; Next-generation Sequencing; Shotgun Metagenomics for Pathogen Detection.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest SM Xie, WJ Chen, JY Zhao, YL Wu, XF Meng, C Ouyang, Z Jiang, ZK Liang, HQ Tan, Y Fang, N Qin, YL Guan, and W Gai were employed by Vision Medicals Center for Infectious Diseases, BGI PathoGenesis Pharmaceutical Technology, Dalian GenTalker Clinical Laboratory, Guangzhou Sagene Biotech Co., Ltd., Guangzhou Kingmed Diagnostics, Hangzhou MatriDx Biotechnology Co., Ltd, Genskey Medical Technology, Co., Ltd., Guangzhou Darui Biotechnology, Co., Ltd., Hangzhou IngeniGen XunMinKang Biotechnology Co., Ltd., Dinfectome Inc, Realbio Genomics Institute, Hugobiotech Co., Ltd., WillingMed Technology (Beijing) Co., Ltd., respectively, outside the submitted work. WL Xing was employed by School of Medicine Tsinghua University and CapitalBio Technology Co., Ltd.
Figures
Similar articles
-
Clinical metagenomics.Nat Rev Genet. 2019 Jun;20(6):341-355. doi: 10.1038/s41576-019-0113-7. Nat Rev Genet. 2019. PMID: 30918369 Free PMC article. Review.
-
Multicenter benchmarking of short and long read wet lab protocols for clinical viral metagenomics.J Clin Virol. 2024 Aug;173:105695. doi: 10.1016/j.jcv.2024.105695. Epub 2024 May 25. J Clin Virol. 2024. PMID: 38823290
-
Limited Correlation of Shotgun Metagenomics Following Host Depletion and Routine Diagnostics for Viruses and Bacteria in Low Concentrated Surrogate and Clinical Samples.Front Cell Infect Microbiol. 2018 Oct 23;8:375. doi: 10.3389/fcimb.2018.00375. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30406048 Free PMC article.
-
From the Pipeline to the Bedside: Advances and Challenges in Clinical Metagenomics.J Infect Dis. 2020 Mar 28;221(Suppl 3):S331-S340. doi: 10.1093/infdis/jiz151. J Infect Dis. 2020. PMID: 31538184 Free PMC article. Review.
-
Species classifier choice is a key consideration when analysing low-complexity food microbiome data.Microbiome. 2018 Mar 20;6(1):50. doi: 10.1186/s40168-018-0437-0. Microbiome. 2018. PMID: 29554948 Free PMC article.
Cited by
-
Performance of mNGS in bronchoalveolar lavage fluid for the diagnosis of invasive pulmonary aspergillosis in non-neutropenic patients.Front Cell Infect Microbiol. 2023 Oct 31;13:1271853. doi: 10.3389/fcimb.2023.1271853. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 38029249 Free PMC article.
-
The effects of sequencing strategies on Metagenomic pathogen detection using bronchoalveolar lavage fluid samples.Heliyon. 2024 Jun 22;10(13):e33429. doi: 10.1016/j.heliyon.2024.e33429. eCollection 2024 Jul 15. Heliyon. 2024. PMID: 39027502 Free PMC article.
-
The performance of metagenomic next-generation sequencing in diagnosing pulmonary infectious diseases using authentic clinical specimens: The Illumina platform versus the Beijing Genomics Institute platform.Front Pharmacol. 2023 Apr 17;14:1164633. doi: 10.3389/fphar.2023.1164633. eCollection 2023. Front Pharmacol. 2023. PMID: 37138853 Free PMC article.
-
Application of metagenomic next-generation sequencing in the diagnosis of urinary tract infection in patients undergoing cutaneous ureterostomy.Front Cell Infect Microbiol. 2023 Jan 27;13:991011. doi: 10.3389/fcimb.2023.991011. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36779185 Free PMC article.
-
Utilizing metagenomic next-generation sequencing for diagnosis and lung microbiome probing of pediatric pneumonia through bronchoalveolar lavage fluid in pediatric intensive care unit: results from a large real-world cohort.Front Cell Infect Microbiol. 2023 Aug 15;13:1200806. doi: 10.3389/fcimb.2023.1200806. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37655299 Free PMC article.
References
-
- Blauwkamp T.A., Thair S., Rosen M.J., et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol. 2019;4(4):663–674. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

