BAP1 forms a trimer with HMGB1 and HDAC1 that modulates gene × environment interaction with asbestos
- PMID: 34815344
- PMCID: PMC8640737
- DOI: 10.1073/pnas.2111946118
BAP1 forms a trimer with HMGB1 and HDAC1 that modulates gene × environment interaction with asbestos
Abstract
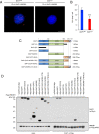
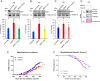
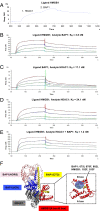
Carriers of heterozygous germline BAP1 mutations (BAP1+/-) are affected by the "BAP1 cancer syndrome." Although they can develop almost any cancer type, they are unusually susceptible to asbestos carcinogenesis and mesothelioma. Here we investigate why among all carcinogens, BAP1 mutations cooperate with asbestos. Asbestos carcinogenesis and mesothelioma have been linked to a chronic inflammatory process promoted by the extracellular release of the high-mobility group box 1 protein (HMGB1). We report that BAP1+/- cells secrete increased amounts of HMGB1, and that BAP1+/- carriers have detectable serum levels of acetylated HMGB1 that further increase when they develop mesothelioma. We linked these findings to our discovery that BAP1 forms a trimeric protein complex with HMGB1 and with histone deacetylase 1 (HDAC1) that modulates HMGB1 acetylation and its release. Reduced BAP1 levels caused increased ubiquitylation and degradation of HDAC1, leading to increased acetylation of HMGB1 and its active secretion that in turn promoted mesothelial cell transformation.
Keywords: HMGB1; asbestos; gene × environment; germline BAP1 mutations; mesothelioma.
Conflict of interest statement
Competing interest statement: M.C. has a patent issued for BAP1. M.C. and H.Y. have two patents issued for HMGB1. M.C. is a board-certified pathologist who provides consultation for pleural pathology, including medical–legal.
Figures




Similar articles
-
Preventive and therapeutic opportunities: targeting BAP1 and/or HMGB1 pathways to diminish the burden of mesothelioma.J Transl Med. 2023 Oct 25;21(1):749. doi: 10.1186/s12967-023-04614-5. J Transl Med. 2023. PMID: 37880686 Free PMC article. Review.
-
Germline mutation of Bap1 accelerates development of asbestos-induced malignant mesothelioma.Cancer Res. 2014 Aug 15;74(16):4388-97. doi: 10.1158/0008-5472.CAN-14-1328. Epub 2014 Jun 13. Cancer Res. 2014. PMID: 24928783 Free PMC article.
-
Minimal asbestos exposure in germline BAP1 heterozygous mice is associated with deregulated inflammatory response and increased risk of mesothelioma.Oncogene. 2016 Apr 14;35(15):1996-2002. doi: 10.1038/onc.2015.243. Epub 2015 Jun 29. Oncogene. 2016. PMID: 26119930 Free PMC article.
-
Germline BAP1 Mutational Landscape of Asbestos-Exposed Malignant Mesothelioma Patients with Family History of Cancer.Cancer Res. 2016 Jan 15;76(2):206-15. doi: 10.1158/0008-5472.CAN-15-0295. Epub 2015 Dec 30. Cancer Res. 2016. PMID: 26719535 Free PMC article.
-
Mesothelioma: Scientific clues for prevention, diagnosis, and therapy.CA Cancer J Clin. 2019 Sep;69(5):402-429. doi: 10.3322/caac.21572. Epub 2019 Jul 8. CA Cancer J Clin. 2019. PMID: 31283845 Free PMC article. Review.
Cited by
-
Identification of TNFRSF1A as a potential biomarker for osteosarcoma.Cancer Biomark. 2024;39(4):299-312. doi: 10.3233/CBM-230086. Cancer Biomark. 2024. PMID: 38250759 Free PMC article.
-
HDAC1: a promising target for cancer treatment: insights from a thorough analysis of tumor functions.Transl Cancer Res. 2024 Oct 31;13(10):5300-5315. doi: 10.21037/tcr-24-23. Epub 2024 Oct 23. Transl Cancer Res. 2024. PMID: 39525004 Free PMC article.
-
HMGB1 as a Key Mediator in Malignant Mesothelioma and a Potential Target for Asbestos-Related Cancer Therapy.Toxics. 2025 May 28;13(6):448. doi: 10.3390/toxics13060448. Toxics. 2025. PMID: 40559922 Free PMC article.
-
Understanding the intersection between placental development and cancer: Lessons from the tumor suppressor BAP1.Commun Biol. 2024 Aug 27;7(1):1053. doi: 10.1038/s42003-024-06689-2. Commun Biol. 2024. PMID: 39191942 Free PMC article. Review.
-
Unraveling Novel Strategies in Mesothelioma Treatments Using a Newly Synthetized Platinum(IV) Compound.Pharmaceutics. 2024 Jul 31;16(8):1015. doi: 10.3390/pharmaceutics16081015. Pharmaceutics. 2024. PMID: 39204360 Free PMC article.
References
-
- Bianchi M. E., et al. , High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol. Rev. 280, 74–82 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

