Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands
- PMID: 34815406
- PMCID: PMC8611045
- DOI: 10.1038/s41467-021-27096-9
Adaptation, spread and transmission of SARS-CoV-2 in farmed minks and associated humans in the Netherlands
Abstract
In the first wave of the COVID-19 pandemic (April 2020), SARS-CoV-2 was detected in farmed minks and genomic sequencing was performed on mink farms and farm personnel. Here, we describe the outbreak and use sequence data with Bayesian phylodynamic methods to explore SARS-CoV-2 transmission in minks and humans on farms. High number of farm infections (68/126) in minks and farm workers (>50% of farms) were detected, with limited community spread. Three of five initial introductions of SARS-CoV-2 led to subsequent spread between mink farms until November 2020. Viruses belonging to the largest cluster acquired an amino acid substitution in the receptor binding domain of the Spike protein (position 486), evolved faster and spread longer and more widely. Movement of people and distance between farms were statistically significant predictors of virus dispersal between farms. Our study provides novel insights into SARS-CoV-2 transmission between mink farms and highlights the importance of combining genetic information with epidemiological information when investigating outbreaks at the animal-human interface.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


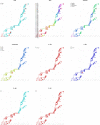

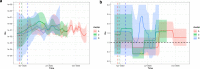

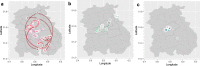
References
-
- El Masry, I. et al. The likelihood of exposure of humans or animals to SARS-CoV-2 from wild, livestock, companion and aquatic animals: qualitative exposure assessment. FAO Animal Production and Health. Paper 181 (FAO, 2020).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

