This is a preprint.
Advancing COVID-19 Diagnosis with Privacy-Preserving Collaboration in Artificial Intelligence
- PMID: 34815983
- PMCID: PMC8609899
Advancing COVID-19 Diagnosis with Privacy-Preserving Collaboration in Artificial Intelligence
Update in
-
Erratum: Author Correction: Advancing COVID-19 diagnosis with privacy-preserving collaboration in artificial intelligence.Nat Mach Intell. 2022;4(4):413. doi: 10.1038/s42256-022-00485-5. Epub 2022 Apr 8. Nat Mach Intell. 2022. PMID: 37520117 Free PMC article.
Abstract
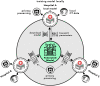
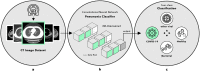
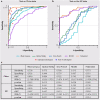
Artificial intelligence (AI) provides a promising substitution for streamlining COVID-19 diagnoses. However, concerns surrounding security and trustworthiness impede the collection of large-scale representative medical data, posing a considerable challenge for training a well-generalised model in clinical practices. To address this, we launch the Unified CT-COVID AI Diagnostic Initiative (UCADI), where the AI model can be distributedly trained and independently executed at each host institution under a federated learning framework (FL) without data sharing. Here we show that our FL model outperformed all the local models by a large yield (test sensitivity /specificity in China: 0.973/0.951, in the UK: 0.730/0.942), achieving comparable performance with a panel of professional radiologists. We further evaluated the model on the hold-out (collected from another two hospitals leaving out the FL) and heterogeneous (acquired with contrast materials) data, provided visual explanations for decisions made by the model, and analysed the trade-offs between the model performance and the communication costs in the federated training process. Our study is based on 9,573 chest computed tomography scans (CTs) from 3,336 patients collected from 23 hospitals located in China and the UK. Collectively, our work advanced the prospects of utilising federated learning for privacy-preserving AI in digital health.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
