This is a preprint.
#COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol
- PMID: 34816263
- PMCID: PMC8609898
- DOI: 10.1101/2021.11.12.468428
#COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy of Delta SARS-CoV-2 in a Respiratory Aerosol
Update in
-
#COVIDisAirborne: AI-enabled multiscale computational microscopy of delta SARS-CoV-2 in a respiratory aerosol.Int J High Perform Comput Appl. 2023 Jan;37(1):28-44. doi: 10.1177/10943420221128233. Epub 2022 Oct 2. Int J High Perform Comput Appl. 2023. PMID: 36647365 Free PMC article.
Abstract
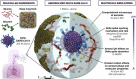
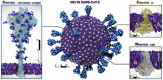
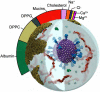
We seek to completely revise current models of airborne transmission of respiratory viruses by providing never-before-seen atomic-level views of the SARS-CoV-2 virus within a respiratory aerosol. Our work dramatically extends the capabilities of multiscale computational microscopy to address the significant gaps that exist in current experimental methods, which are limited in their ability to interrogate aerosols at the atomic/molecular level and thus ob-scure our understanding of airborne transmission. We demonstrate how our integrated data-driven platform provides a new way of exploring the composition, structure, and dynamics of aerosols and aerosolized viruses, while driving simulation method development along several important axes. We present a series of initial scientific discoveries for the SARS-CoV-2 Delta variant, noting that the full scientific impact of this work has yet to be realized.
Acm reference format: Abigail Dommer 1† , Lorenzo Casalino 1† , Fiona Kearns 1† , Mia Rosenfeld 1 , Nicholas Wauer 1 , Surl-Hee Ahn 1 , John Russo, 2 Sofia Oliveira 3 , Clare Morris 1 , AnthonyBogetti 4 , AndaTrifan 5,6 , Alexander Brace 5,7 , TerraSztain 1,8 , Austin Clyde 5,7 , Heng Ma 5 , Chakra Chennubhotla 4 , Hyungro Lee 9 , Matteo Turilli 9 , Syma Khalid 10 , Teresa Tamayo-Mendoza 11 , Matthew Welborn 11 , Anders Christensen 11 , Daniel G. A. Smith 11 , Zhuoran Qiao 12 , Sai Krishna Sirumalla 11 , Michael O'Connor 11 , Frederick Manby 11 , Anima Anandkumar 12,13 , David Hardy 6 , James Phillips 6 , Abraham Stern 13 , Josh Romero 13 , David Clark 13 , Mitchell Dorrell 14 , Tom Maiden 14 , Lei Huang 15 , John McCalpin 15 , Christo- pherWoods 3 , Alan Gray 13 , MattWilliams 3 , Bryan Barker 16 , HarindaRajapaksha 16 , Richard Pitts 16 , Tom Gibbs 13 , John Stone 6 , Daniel Zuckerman 2 *, Adrian Mulholland 3 *, Thomas MillerIII 11,12 *, ShantenuJha 9 *, Arvind Ramanathan 5 *, Lillian Chong 4 *, Rommie Amaro 1 *. 2021. #COVIDisAirborne: AI-Enabled Multiscale Computational Microscopy ofDeltaSARS-CoV-2 in a Respiratory Aerosol. In Supercomputing '21: International Conference for High Perfor-mance Computing, Networking, Storage, and Analysis . ACM, New York, NY, USA, 14 pages. https://doi.org/finalDOI.
Figures



References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., and Lindahl E.. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX, 1–2:19–25, 2015. ISSN 2352–7110. doi: 10.1016/j.softx.2015.06.001. URL http://www.sciencedirect.com/science/article/pii/S2352711015000059. - DOI
-
- Bangaru S., Ozorowski G., Turner H. L., Antanasijevic A., Huang D., Wang X., Torres J. L., Diedrich J. K., Tian J.-H., Portnoff A. D., Patel N., Massare M. J., Yates J. R., Nemazee D., Paulson J. C., Glenn G., Smith G., and Ward A. B.. Structural analysis of full-length sars-cov-2 spike protein from an advanced vaccine candidate. Science, 370(6520):1089–1094, 2020. doi: 10.1126/science.abe1502. - DOI - PMC - PubMed
-
- Beglov D. and Roux B.. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. The Journal of Chemical Physics, 1994. ISSN 00219606. doi: 10.1063/1.466711. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
