Blood stem cell PU.1 upregulation is a consequence of differentiation without fast autoregulation
- PMID: 34817548
- PMCID: PMC8624737
- DOI: 10.1084/jem.20202490
Blood stem cell PU.1 upregulation is a consequence of differentiation without fast autoregulation
Abstract
Transcription factors (TFs) regulate cell fates, and their expression must be tightly regulated. Autoregulation is assumed to regulate many TFs' own expression to control cell fates. Here, we manipulate and quantify the (auto)regulation of PU.1, a TF controlling hematopoietic stem and progenitor cells (HSPCs), and correlate it to their future fates. We generate transgenic mice allowing both inducible activation of PU.1 and noninvasive quantification of endogenous PU.1 protein expression. The quantified HSPC PU.1 dynamics show that PU.1 up-regulation occurs as a consequence of hematopoietic differentiation independently of direct fast autoregulation. In contrast, inflammatory signaling induces fast PU.1 up-regulation, which does not require PU.1 expression or its binding to its own autoregulatory enhancer. However, the increased PU.1 levels induced by inflammatory signaling cannot be sustained via autoregulation after removal of the signaling stimulus. We conclude that PU.1 overexpression induces HSC differentiation before PU.1 up-regulation, only later generating cell types with intrinsically higher PU.1.
© 2021 Ahmed et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures

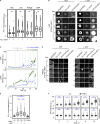
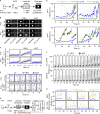
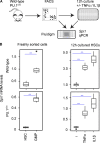
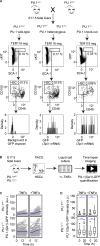


References
-
- Bouchoucha, Y.X., Reingruber J., Labalette C., Wassef M.A., Thierion E., Desmarquet-Trin Dinh C., Holcman D., Gilardi-Hebenstreit P., and Charnay P.. 2013. Dissection of a Krox20 positive feedback loop driving cell fate choices in hindbrain patterning. Mol. Syst. Biol. 9:690. 10.1038/msb.2013.46 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

