Site-Specific Labeling of Endogenous Proteins Using CoLDR Chemistry
- PMID: 34817989
- PMCID: PMC8662641
- DOI: 10.1021/jacs.1c06167
Site-Specific Labeling of Endogenous Proteins Using CoLDR Chemistry
Abstract
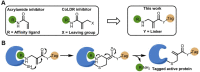
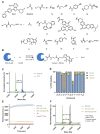
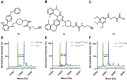
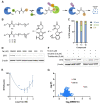
Chemical modifications of native proteins can affect their stability, activity, interactions, localization, and more. However, there are few nongenetic methods for the installation of chemical modifications at a specific protein site in cells. Here we report a covalent ligand directed release (CoLDR) site-specific labeling strategy, which enables the installation of a variety of functional tags on a target protein while releasing the directing ligand. Using this approach, we were able to label various proteins such as BTK, K-RasG12C, and SARS-CoV-2 PLpro with different tags. For BTK we have shown selective labeling in cells of both alkyne and fluorophores tags. Protein labeling by traditional affinity methods often inhibits protein activity since the directing ligand permanently occupies the target binding pocket. We have shown that using CoLDR chemistry, modification of BTK by these probes in cells preserves its activity. We demonstrated several applications for this approach including determining the half-life of BTK in its native environment with minimal perturbation, as well as quantification of BTK degradation by a noncovalent proteolysis targeting chimera (PROTAC) by in-gel fluorescence. Using an environment-sensitive "turn-on" fluorescent probe, we were able to monitor ligand binding to the active site of BTK. Finally, we have demonstrated efficient CoLDR-based BTK PROTACs (DC50 < 100 nM), which installed a CRBN binder onto BTK. This approach joins very few available labeling strategies that maintain the target protein activity and thus makes an important addition to the toolbox of chemical biology.
Conflict of interest statement
The authors declare the following competing financial interest(s): N.L., R.N.R., A.R., and E.R. are inventors on a patent application describing this technology.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

