ER-localized phosphatidylethanolamine synthase plays a conserved role in lipid droplet formation
- PMID: 34818062
- PMCID: PMC8886813
- DOI: 10.1091/mbc.E21-11-0558-T
ER-localized phosphatidylethanolamine synthase plays a conserved role in lipid droplet formation
Abstract
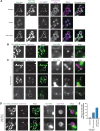
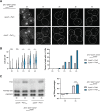
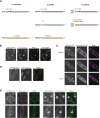
The asymmetric distribution of phospholipids in membranes is a fundamental principle of cellular compartmentalization and organization. Phosphatidylethanolamine (PE), a nonbilayer phospholipid that contributes to organelle shape and function, is synthesized at several subcellular localizations via semiredundant pathways. Previously, we demonstrated in budding yeast that the PE synthase Psd1, which primarily operates on the mitochondrial inner membrane, is additionally targeted to the ER. While ER-localized Psd1 is required to support cellular growth in the absence of redundant pathways, its physiological function is unclear. We now demonstrate that ER-localized Psd1 sublocalizes on the ER to lipid droplet (LD) attachment sites and show it is specifically required for normal LD formation. We also find that the role of phosphatidylserine decarboxylase (PSD) enzymes in LD formation is conserved in other organisms. Thus we have identified PSD enzymes as novel regulators of LDs and demonstrate that both mitochondria and LDs in yeast are organized and shaped by the spatial positioning of a single PE synthesis enzyme.
Figures




References
-
- Arguello-Miranda O, Liu Y, Wood NE, Kositangool P, Doncic A (2018). Integration of multiple metabolic signals determines cell fate prior to commitment. Mol Cell. 71, 733–744.e711. - PubMed
-
- Ben M’barek K, Ajjaji D, Chorlay A, Vanni S, Foret L, Thiam AR (2017). ER Membrane phospholipids and surface tension control cellular lipid droplet formation. Dev Cell 41, 591–604.e597. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

