The ARF GAPs ELMOD1 and ELMOD3 act at the Golgi and cilia to regulate ciliogenesis and ciliary protein traffic
- PMID: 34818063
- PMCID: PMC9236152
- DOI: 10.1091/mbc.E21-09-0443
The ARF GAPs ELMOD1 and ELMOD3 act at the Golgi and cilia to regulate ciliogenesis and ciliary protein traffic
Erratum in
-
The ARF GAPs ELMOD1 and ELMOD3 act at the Golgi and cilia to regulate ciliogenesis and ciliary protein traffic.Mol Biol Cell. 2022 Jun 1;33(7):cor1. doi: 10.1091/mbc.E21-09-0443_corr. Mol Biol Cell. 2022. PMID: 35612986 Free PMC article. No abstract available.
Abstract
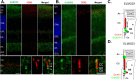
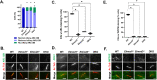
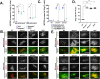
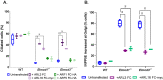
ELMODs are a family of three mammalian paralogues that display GTPase-activating protein (GAP) activity toward a uniquely broad array of ADP-ribosylation factor (ARF) family GTPases that includes ARF-like (ARL) proteins. ELMODs are ubiquitously expressed in mammalian tissues, highly conserved across eukaryotes, and ancient in origin, being present in the last eukaryotic common ancestor. We described functions of ELMOD2 in immortalized mouse embryonic fibroblasts (MEFs) in the regulation of cell division, microtubules, ciliogenesis, and mitochondrial fusion. Here, using similar strategies with the paralogues ELMOD1 and ELMOD3, we identify novel functions and locations of these cell regulators and compare them to those of ELMOD2, allowing the determination of functional redundancy among the family members. We found strong similarities in phenotypes resulting from deletion of either Elmod1 or Elmod3 and marked differences from those arising in Elmod2 deletion lines. Deletion of either Elmod1 or Elmod3 results in the decreased ability of cells to form primary cilia, loss of a subset of proteins from cilia, and accumulation of some ciliary proteins at the Golgi, predicted to result from compromised traffic from the Golgi to cilia. These phenotypes are reversed upon activating mutant expression of either ARL3 or ARL16, linking their roles to ELMOD1/3 actions.
Figures


References
-
- Bielas SL, Silhavy JL, Brancati F, Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS, Abdel-Aleem A, Rosti RO, et al. (2009). Mutations in INPP5E, encoding inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol signaling to the ciliopathies. Nat Genet 41, 1032–1036. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

