Type IX Secretion System Effectors and Virulence of the Model Flavobacterium columnare Strain MS-FC-4
- PMID: 34818105
- PMCID: PMC8824203
- DOI: 10.1128/AEM.01705-21
Type IX Secretion System Effectors and Virulence of the Model Flavobacterium columnare Strain MS-FC-4
Abstract
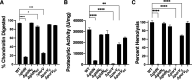
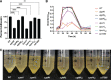
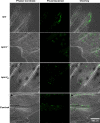
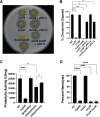
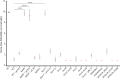
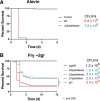
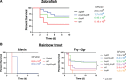
Flavobacterium columnare causes columnaris disease in wild and cultured freshwater fish and is a major problem for sustainable aquaculture worldwide. The F. columnare type IX secretion system (T9SS) secretes many proteins and is required for virulence. The T9SS component GldN is required for secretion and gliding motility over surfaces. Genetic manipulation of F. columnare is inefficient, which has impeded identification of secreted proteins that are critical for virulence. Here, we identified a virulent wild-type F. columnare strain (MS-FC-4) that is highly amenable to genetic manipulation. This facilitated isolation and characterization of two deletion mutants lacking core components of the T9SS. Deletion of gldN disrupted protein secretion and gliding motility and eliminated virulence in zebrafish and rainbow trout. Deletion of porV disrupted secretion and virulence but not motility. Both mutants exhibited decreased extracellular proteolytic, hemolytic, and chondroitin sulfate lyase activities. They also exhibited decreased biofilm formation and decreased attachment to fish fins and other surfaces. Using genomic and proteomic approaches, we identified proteins secreted by the T9SS. We deleted 10 genes encoding secreted proteins and characterized the virulence of mutants lacking individual or multiple secreted proteins. A mutant lacking two genes encoding predicted peptidases exhibited reduced virulence in rainbow trout, and mutants lacking a predicted cytolysin showed reduced virulence in zebrafish and rainbow trout. The results establish F. columnare strain MS-FC-4 as a genetically amenable model to identify virulence factors. This may aid development of measures to control columnaris disease and impact fish health and sustainable aquaculture. IMPORTANCE Flavobacterium columnare causes columnaris disease in wild and aquaculture-reared freshwater fish and is a major problem for aquaculture. Little is known regarding the virulence factors involved in this disease, and control measures are inadequate. The type IX secretion system (T9SS) secretes many proteins and is required for virulence, but the secreted virulence factors are not known. We identified a strain of F. columnare (MS-FC-4) that is well suited for genetic manipulation. The components of the T9SS and the proteins secreted by this system were identified. Deletion of core T9SS genes eliminated virulence. Genes encoding 10 secreted proteins were deleted. Deletion of two peptidase-encoding genes resulted in decreased virulence in rainbow trout, and deletion of a cytolysin-encoding gene resulted in decreased virulence in rainbow trout and zebrafish. Secreted peptidases and cytolysins are likely virulence factors and are targets for the development of control measures.
Keywords: columnaris disease; fish pathogen; flavobacterium; type IX secretion system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Secreted peptidases contribute to virulence of fish pathogen Flavobacterium columnare.Front Cell Infect Microbiol. 2023 Feb 3;13:1093393. doi: 10.3389/fcimb.2023.1093393. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36816589 Free PMC article.
-
The Type IX Secretion System Is Required for Virulence of the Fish Pathogen Flavobacterium columnare.Appl Environ Microbiol. 2017 Nov 16;83(23):e01769-17. doi: 10.1128/AEM.01769-17. Print 2017 Dec 1. Appl Environ Microbiol. 2017. PMID: 28939608 Free PMC article.
-
Gliding motility proteins GldJ and SprB contribute to Flavobacterium columnare virulence.J Bacteriol. 2024 Apr 18;206(4):e0006824. doi: 10.1128/jb.00068-24. Epub 2024 Mar 22. J Bacteriol. 2024. PMID: 38517170 Free PMC article.
-
Columnaris disease in fish: a review with emphasis on bacterium-host interactions.Vet Res. 2013 Apr 24;44(1):27. doi: 10.1186/1297-9716-44-27. Vet Res. 2013. PMID: 23617544 Free PMC article. Review.
-
The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function.Front Cell Infect Microbiol. 2017 May 26;7:215. doi: 10.3389/fcimb.2017.00215. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28603700 Free PMC article. Review.
Cited by
-
Draft genome sequence of the emerging fish pathogen Flavobacterium tructae strain S12.Microbiol Resour Announc. 2023 Dec 14;12(12):e0048823. doi: 10.1128/MRA.00488-23. Epub 2023 Nov 3. Microbiol Resour Announc. 2023. PMID: 37921484 Free PMC article.
-
A tradeoff between bacteriophage resistance and bacterial motility is mediated by the Rcs phosphorelay in Escherichia coli.Microbiology (Reading). 2024 Aug;170(8):001491. doi: 10.1099/mic.0.001491. Microbiology (Reading). 2024. PMID: 39194382 Free PMC article.
-
Opportunistic diseases in marine eukaryotes: Could Bacteroidota be the next threat to ocean life?Environ Microbiol. 2022 Oct;24(10):4505-4518. doi: 10.1111/1462-2920.16094. Epub 2022 Aug 4. Environ Microbiol. 2022. PMID: 35706128 Free PMC article.
-
Secreted peptidases contribute to virulence of fish pathogen Flavobacterium columnare.Front Cell Infect Microbiol. 2023 Feb 3;13:1093393. doi: 10.3389/fcimb.2023.1093393. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36816589 Free PMC article.
-
Secreted Extracellular Products of Flavobacterium covae as Potential Immunogenic Factors for Protection against Columnaris Disease in Channel Catfish (Ictalurus punctatus).Pathogens. 2023 Jun 7;12(6):808. doi: 10.3390/pathogens12060808. Pathogens. 2023. PMID: 37375498 Free PMC article.
References
-
- Davis HS. 1922. A new bacterial disease of freshwater fishes. Bull US Bureau Fish 38:261–280.
-
- Bullock GL, Hsu TC, Shotts EB. 1986. Columnaris disease of fishes. Fish Disease Leaflet 72. U.S. Fish and Wildlife Service, U.S. Department of the Interior, Washington, DC.
Publication types
MeSH terms
Supplementary concepts
LinkOut - more resources
Full Text Sources

