Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance
- PMID: 34818780
- PMCID: PMC8386095
- DOI: 10.1016/j.scitotenv.2021.149877
Minimizing errors in RT-PCR detection and quantification of SARS-CoV-2 RNA for wastewater surveillance
Abstract
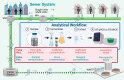
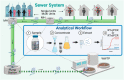
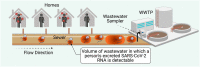
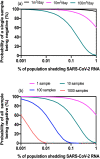
Wastewater surveillance for pathogens using reverse transcription-polymerase chain reaction (RT-PCR) is an effective and resource-efficient tool for gathering community-level public health information, including the incidence of coronavirus disease-19 (COVID-19). Surveillance of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) in wastewater can potentially provide an early warning signal of COVID-19 infections in a community. The capacity of the world's environmental microbiology and virology laboratories for SARS-CoV-2 RNA characterization in wastewater is increasing rapidly. However, there are no standardized protocols or harmonized quality assurance and quality control (QA/QC) procedures for SARS-CoV-2 wastewater surveillance. This paper is a technical review of factors that can cause false-positive and false-negative errors in the surveillance of SARS-CoV-2 RNA in wastewater, culminating in recommended strategies that can be implemented to identify and mitigate some of these errors. Recommendations include stringent QA/QC measures, representative sampling approaches, effective virus concentration and efficient RNA extraction, PCR inhibition assessment, inclusion of sample processing controls, and considerations for RT-PCR assay selection and data interpretation. Clear data interpretation guidelines (e.g., determination of positive and negative samples) are critical, particularly when the incidence of SARS-CoV-2 in wastewater is low. Corrective and confirmatory actions must be in place for inconclusive results or results diverging from current trends (e.g., initial onset or reemergence of COVID-19 in a community). It is also prudent to perform interlaboratory comparisons to ensure results' reliability and interpretability for prospective and retrospective analyses. The strategies that are recommended in this review aim to improve SARS-CoV-2 characterization and detection for wastewater surveillance applications. A silver lining of the COVID-19 pandemic is that the efficacy of wastewater surveillance continues to be demonstrated during this global crisis. In the future, wastewater should also play an important role in the surveillance of a range of other communicable diseases.
Keywords: COVID-19; False negative; False positive; RT-PCR; SARS-CoV-2; Surveillance; Wastewater.
Crown Copyright © 2021. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
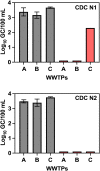
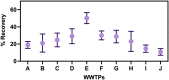
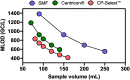
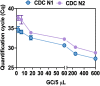
References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., Brien J.W.O., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., Gyawali P., Korajkic A., Riddell S., Sherchan S.P., Simpson S.L., Sirikanchana K., Symonds E.M., Verhagen R., Vasan S.S., Kitajima M., Bivins A. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 - PMC - PubMed
-
- Ahmed W., Bivins A., Bertsch P.M., Bibby K., Choi P.M., Farkas K., Gyawali P., Hamilton K.A., Haramoto E., Kitajima M., Simpson S.L., Tandukar S., Thomas K.V., Mueller J.F. Surveillance of SARS-CoV-2 RNA in wastewater: methods optimization and quality control are crucial for generating reliable public health information. Curr. Opin. Environ. Sci. Health. 2020;17:82–93. - PMC - PubMed
-
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., Haramoto E., Gyawali P., Korajkic A., McMinn B.R., Mueller J.F., Simpson S.L., Smith W.J.M., Symonds E.M., Thomas K.V., Verhagen R., Kitajima M. Comparison of virus concentration methods for the RT-qPCR based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

