A study of generalization and compatibility performance of 3D U-Net segmentation on multiple heterogeneous liver CT datasets
- PMID: 34819022
- PMCID: PMC8611902
- DOI: 10.1186/s12880-021-00708-y
A study of generalization and compatibility performance of 3D U-Net segmentation on multiple heterogeneous liver CT datasets
Abstract
Background: Most existing algorithms have been focused on the segmentation from several public Liver CT datasets scanned regularly (no pneumoperitoneum and horizontal supine position). This study primarily segmented datasets with unconventional liver shapes and intensities deduced by contrast phases, irregular scanning conditions, different scanning objects of pigs and patients with large pathological tumors, which formed the multiple heterogeneity of datasets used in this study.
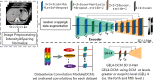
Methods: The multiple heterogeneous datasets used in this paper includes: (1) One public contrast-enhanced CT dataset and one public non-contrast CT dataset; (2) A contrast-enhanced dataset that has abnormal liver shape with very long left liver lobes and large-sized liver tumors with abnormal presets deduced by microvascular invasion; (3) One artificial pneumoperitoneum dataset under the pneumoperitoneum and three scanning profiles (horizontal/left/right recumbent position); (4) Two porcine datasets of Bama type and domestic type that contains pneumoperitoneum cases but with large anatomy discrepancy with humans. The study aimed to investigate the segmentation performances of 3D U-Net in: (1) generalization ability between multiple heterogeneous datasets by cross-testing experiments; (2) the compatibility when hybrid training all datasets in different sampling and encoder layer sharing schema. We further investigated the compatibility of encoder level by setting separate level for each dataset (i.e., dataset-wise convolutions) while sharing the decoder.
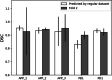
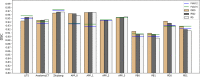
Results: Model trained on different datasets has different segmentation performance. The prediction accuracy between LiTS dataset and Zhujiang dataset was about 0.955 and 0.958 which shows their good generalization ability due to that they were all contrast-enhanced clinical patient datasets scanned regularly. For the datasets scanned under pneumoperitoneum, their corresponding datasets scanned without pneumoperitoneum showed good generalization ability. Dataset-wise convolution module in high-level can improve the dataset unbalance problem. The experimental results will facilitate researchers making solutions when segmenting those special datasets.
Conclusions: (1) Regularly scanned datasets is well generalized to irregularly ones. (2) The hybrid training is beneficial but the dataset imbalance problem always exits due to the multi-domain homogeneity. The higher levels encoded more domain specific information than lower levels and thus were less compatible in terms of our datasets.
Keywords: Dataset-wise convolution; Generalization; Liver segmentation; U-Net.
© 2021. The Author(s).
Conflict of interest statement
The authors have no competing interest to declare.
Figures




References
-
- Moghbel M, Mashohor S, Mahmud R, Saripan MIB. Review of liver segmentation and computer assisted detection/diagnosis methods in computed tomography. Artif Intell Rev. 2017;50(4):497–537. doi: 10.1007/s10462-017-9550-x. - DOI
-
- Li C, Wang X, Eberl S, Fulham M, Yin Y, Feng D. Fully automated liver segmentation for low-and high-contrast CT volumes based on probabilistic atlases. In: Proceedings of the ICIP; 2010. p. 1733–6.
-
- Massoptier L, Casciaro S. Fully automatic liver segmentation through graph-cut technique. In: Proceedings of the IEMBS, Lyon; 2007. p. 5243–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

