High efficiency closed-system gene transfer using automated spinoculation
- PMID: 34819105
- PMCID: PMC8675485
- DOI: 10.1186/s12967-021-03126-4
High efficiency closed-system gene transfer using automated spinoculation
Abstract
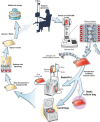
Background: Gene transfer is an important tool for cellular therapies. Lentiviral vectors are most effectively transferred into lymphocytes or hematopoietic progenitor cells using spinoculation. To enable cGMP (current Good Manufacturing Practice)-compliant cell therapy production, we developed and compared a closed-system spinoculation method that uses cell culture bags, and an automated closed system spinoculation method to decrease technician hands on time and reduce the likelihood for microbial contamination.
Methods: Sepax spinoculation, bag spinoculation, and static bag transduction without spinoculation were compared for lentiviral gene transfer in lymphocytes collected by apheresis. The lymphocytes were transduced once and cultured for 9 days. The lentiviral vectors tested encoded a CD19/CD22 Bispecific Chimeric Antigen Receptor (CAR), a FGFR4-CAR, or a CD22-CAR. Sepax spinoculation times were evaluated by testing against bag spinoculation and static transduction to optimize the Sepax spin time. The Sepax spinoculation was then used to test the transduction of different CAR vectors. The performance of the process using healthy donor and a patient sample was evaluated. Functional assessment was performed of the CD19/22 and CD22 CAR T-cells using killing assays against the NALM6 tumor cell line and cytokine secretion analysis. Finally, gene expression of the transduced T-cells was examined to determine if there were any major changes that may have occurred as a result of the spinoculation process.
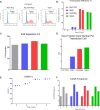
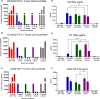
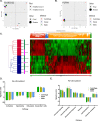
Results: The process of spinoculation lead to significant enhancement in gene transfer. Sepax spinoculation using a 1-h spin time showed comparable transduction efficiency to the bag spinoculation, and much greater than the static bag transduction method (83.4%, 72.8%, 35.7% n = 3). The performance of three different methods were consistent for all lentiviral vectors tested and no significant difference was observed when using starting cells from healthy donor versus a patient sample. Sepax spinoculation does not affect the function of the CAR T-cells against tumor cells, as these cells appeared to kill target cells equally well. Spinoculation also does not appear to affect gene expression patterns that are necessary for imparting function on the cell.
Conclusions: Closed system-bag spinoculation resulted in more efficient lymphocyte gene transfer than standard bag transductions without spinoculation. This method is effective for both retroviral and lentiviral vector gene transfer in lymphocytes and may be a feasible approach for gene transfer into other cell types including hematopoietic and myeloid progenitors. Sepax spinoculation further improved upon the process by offering an automated, closed system approach that significantly decreased hands-on time while also decreasing the risk of culture bag tears and microbial contamination.
Keywords: CAR T-cell; Gene transfer; Sepax; Spinoculation.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. 2015;385(9967):517–528. doi: 10.1016/S0140-6736(14)61403-3. - DOI - PMC - PubMed
-
- Kochenderfer JN, Dudley ME, Kassim SH, Somerville RP, Carpenter RO, Stetler-Stevenson M, et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol. 2015;33(6):540–549. doi: 10.1200/JCO.2014.56.2025. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

