Identification of a novel prognosis-associated ceRNA network in lung adenocarcinoma via bioinformatics analysis
- PMID: 34819106
- PMCID: PMC8611860
- DOI: 10.1186/s12938-021-00952-x
Identification of a novel prognosis-associated ceRNA network in lung adenocarcinoma via bioinformatics analysis
Abstract
Background: Lung adenocarcinoma (LUAD) is the most common subtype of nonsmall-cell lung cancer (NSCLC) and has a high incidence rate and mortality. The survival of LUAD patients has increased with the development of targeted therapeutics, but the prognosis of these patients is still poor. Long noncoding RNAs (lncRNAs) play an important role in the occurrence and development of LUAD. The purpose of this study was to identify novel abnormally regulated lncRNA-microRNA (miRNA)-messenger RNA (mRNA) competing endogenous RNA (ceRNA) networks that may suggest new therapeutic targets for LUAD or relate to LUAD prognosis.
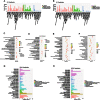
Methods: We used the SBC human ceRNA array V1.0 to screen for differentially expressed (DE) lncRNAs and mRNAs in four paired LUAD samples. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed to annotate the DE lncRNAs and mRNAs. R bioinformatics packages, The Cancer Genome Atlas (TCGA) LUAD database, and Kaplan-Meier (KM) survival analysis tools were used to validate the microarray data and construct the lncRNA-miRNA-mRNA ceRNA regulatory network. Then, quantitative real-time PCR (qRT-PCR) was used to validate the DE lncRNAs in 7 LUAD cell lines.
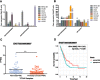
Results: A total of 2819 DE lncRNAs and 2396 DE mRNAs (P < 0.05 and fold change ≥ 2 or ≤ 0.5) were identified in four paired LUAD tissue samples. In total, 255 of the DE lncRNAs were also identified in TCGA. The GO and KEGG analysis results suggested that the DE genes were most enriched in angiogenesis and cell proliferation, and were closely related to human cancers. Moreover, the differential expression of ENST00000609697, ENST00000602992, and NR_024321 was consistent with the microarray data, as determined by qRT-PCR validation in 7 LUAD cell lines; however, only ENST00000609697 was associated with the overall survival of LUAD patients (log-rank P = 0.029). Finally, through analysis of ENST00000609697 target genes, we identified the ENST00000609697-hsa-miR-6791-5p-RASL12 ceRNA network, which may play a tumor-suppressive role in LUAD.
Conclusion: ENST00000609697 was abnormally expressed in LUAD. Furthermore, downregulation of ENST00000609697 and its target gene RASL12 was associated with poor prognosis in LUAD. The ENST00000609697-hsa-miR-6791-5p-RASL12 axis may play a tumor-suppressive role. These results suggest new potential prognostic and therapeutic biomarkers for LUAD.
Keywords: Biomarker; CeRNA; LncRNAs; Lung adenocarcinoma; Prognosis.
© 2021. The Author(s).
Conflict of interest statement
All authors declare that there are no conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
- 2017Q002/The Youth Scientific Research Fund Project of the Affiliated Hospital of Hebei University
- 2015Z005/The Key Scientific Research Fund Project of the Affiliated Hospital of Hebei University
- 361007/The Hebei Province Government Foundation for Clinical Medical Talent Training and Basic Research Projects
- 605020521007/The Outstanding Young Scientific Research and Innovation Team of Hebei University
LinkOut - more resources
Full Text Sources
Medical

