Live-Cell Imaging Shows Uneven Segregation of Extrachromosomal DNA Elements and Transcriptionally Active Extrachromosomal DNA Hubs in Cancer
- PMID: 34819316
- PMCID: PMC8831456
- DOI: 10.1158/2159-8290.CD-21-1376
Live-Cell Imaging Shows Uneven Segregation of Extrachromosomal DNA Elements and Transcriptionally Active Extrachromosomal DNA Hubs in Cancer
Abstract
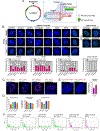
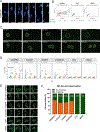
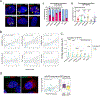
Oncogenic extrachromosomal DNA elements (ecDNA) play an important role in tumor evolution, but our understanding of ecDNA biology is limited. We determined the distribution of single-cell ecDNA copy number across patient tissues and cell line models and observed how cell-to-cell ecDNA frequency varies greatly. The exceptional intratumoral heterogeneity of ecDNA suggested ecDNA-specific replication and propagation mechanisms. To evaluate the transfer of ecDNA genetic material from parental to offspring cells during mitosis, we established the CRISPR-based ecTag method. ecTag leverages ecDNA-specific breakpoint sequences to tag ecDNA with fluorescent markers in living cells. Applying ecTag during mitosis revealed disjointed ecDNA inheritance patterns, enabling rapid ecDNA accumulation in individual cells. After mitosis, ecDNAs clustered into ecDNA hubs, and ecDNA hubs colocalized with RNA polymerase II, promoting transcription of cargo oncogenes. Our observations provide direct evidence for uneven segregation of ecDNA and shed new light on mechanisms through which ecDNAs contribute to oncogenesis. SIGNIFICANCE: ecDNAs are vehicles for oncogene amplification. The circular nature of ecDNA affords unique properties, such as mobility and ecDNA-specific replication and segregation behavior. We uncovered fundamental ecDNA properties by tracking ecDNAs in live cells, highlighting uneven and random segregation and ecDNA hubs that drive cargo gene transcription.See related commentary by Henssen, p. 293.This article is highlighted in the In This Issue feature, p. 275.
©2021 American Association for Cancer Research.
Figures

Comment in
-
ecDNAs personify cancer gangsters.Mol Cell. 2022 Feb 3;82(3):500-502. doi: 10.1016/j.molcel.2022.01.003. Mol Cell. 2022. PMID: 35120647
-
Shedding Light on ecDNA Behavior Using CRISPR-Based Live-Cell Imaging.Cancer Discov. 2022 Feb;12(2):293-295. doi: 10.1158/2159-8290.CD-21-1525. Cancer Discov. 2022. PMID: 35140176
References
-
- Amirouchene-Angelozzi N, Swanton C, Bardelli A. Tumor Evolution as a Therapeutic Target. Cancer discovery 2017;7:805–817 doi 10.1158/2159-8290.cd-17-0343. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

