ATR inhibition enables complete tumour regression in ALK-driven NB mouse models
- PMID: 34819497
- PMCID: PMC8613282
- DOI: 10.1038/s41467-021-27057-2
ATR inhibition enables complete tumour regression in ALK-driven NB mouse models
Abstract
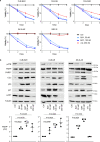
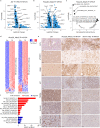
High-risk neuroblastoma (NB) often involves MYCN amplification as well as mutations in ALK. Currently, high-risk NB presents significant clinical challenges, and additional therapeutic options are needed. Oncogenes like MYCN and ALK result in increased replication stress in cancer cells, offering therapeutically exploitable options. We have pursued phosphoproteomic analyses highlighting ATR activity in ALK-driven NB cells, identifying the BAY1895344 ATR inhibitor as a potent inhibitor of NB cell growth and proliferation. Using RNA-Seq, proteomics and phosphoproteomics we characterize NB cell and tumour responses to ATR inhibition, identifying key components of the DNA damage response as ATR targets in NB cells. ATR inhibition also produces robust responses in mouse models. Remarkably, a 2-week combined ATR/ALK inhibition protocol leads to complete tumor regression in two independent genetically modified mouse NB models. These results suggest that NB patients, particularly in high-risk groups with oncogene-induced replication stress, may benefit from ATR inhibition as therapeutic intervention.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Matthay KK, et al. Neuroblastoma. Nat. Rev. Dis. Prim. 2016;2:16078. - PubMed
-
- Ladenstein R, et al. Busulfan and melphalan versus carboplatin, etoposide, and melphalan as high-dose chemotherapy for high-risk neuroblastoma (HR-NBL1/SIOPEN): an international, randomised, multi-arm, open-label, phase 3 trial. Lancet Oncol. 2017;18:500–514. - PubMed
-
- De Brouwer S, et al. Meta-analysis of neuroblastomas reveals a skewed ALK mutation spectrum in tumors with MYCN amplification. Clin. Cancer Res. 2010;16:4353–4362. - PubMed
-
- Grobner SN, et al. The landscape of genomic alterations across childhood cancers. Nature. 2018;555:321–327. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

